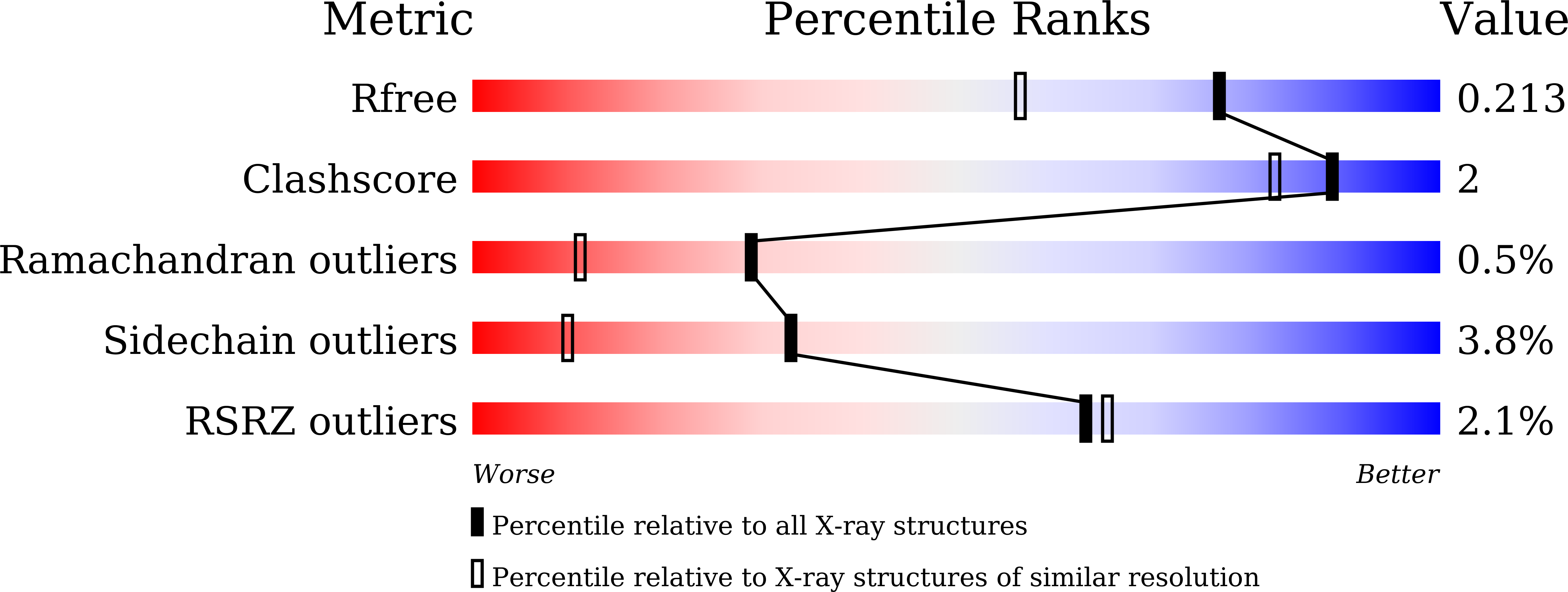
Trypanosomal Phosphodiesterase B1 and B2 as a Potential Therapy for Human African Trypanosomiasis
Ng, P.S., Noble, C.G., Ng, F.S.B., Chew, S.H., Lim, C.C., Manoharan, V., Wan, K.F., Vachaspati, P., Kaiser, M., Ma, N.L., Gedeck, P., Kounde, C., Rao, S.P.S.To be published.