A Second RNA-Binding Site in the NS1 Protein of Influenza B Virus.
Ma, L.C., Guan, R., Hamilton, K., Aramini, J.M., Mao, L., Wang, S., Krug, R.M., Montelione, G.T.(2016) Structure 24: 1562-1572
- PubMed: 27545620
- DOI: https://doi.org/10.1016/j.str.2016.07.001
- Primary Citation of Related Structures:
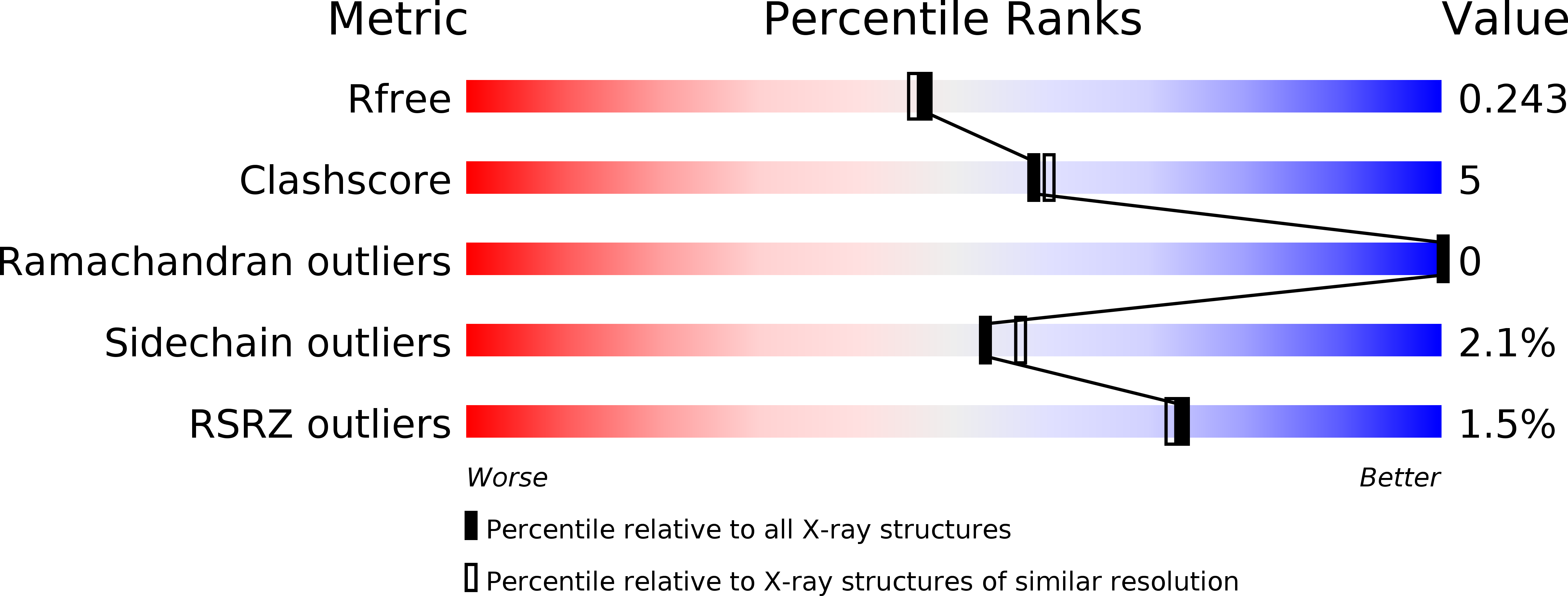
5DIL - PubMed Abstract:
Influenza viruses cause a highly contagious respiratory disease in humans. The NS1 proteins of influenza A and B viruses (NS1A and NS1B proteins, respectively) are composed of two domains, a dimeric N-terminal domain and a C-terminal domain, connected by a flexible polypeptide linker. Here we report the 2.0-Å X-ray crystal structure and nuclear magnetic resonance studies of the NS1B C-terminal domain, which reveal a novel and unexpected basic RNA-binding site that is not present in the NS1A protein. We demonstrate that single-site alanine replacements of basic residues in this site lead to reduced RNA-binding activity, and that recombinant influenza B viruses expressing these mutant NS1B proteins are severely attenuated in replication. This novel RNA-binding site of NS1B is required for optimal influenza B virus replication. Most importantly, this study reveals an unexpected RNA-binding function in the C-terminal domain of NS1B, a novel function that distinguishes influenza B viruses from influenza A viruses.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, Center for Advanced Biotechnology and Medicine, Rutgers, The State University of New Jersey, Northeast Structural Genomics Consortium, Piscataway, NJ 08854, USA.