Structural analysis of a function-associated loop mutant of the substrate-recognition domain of Fbs1 ubiquitin ligase
Nishio, K., Yoshida, Y., Tanaka, K., Mizushima, T.(2016) Acta Crystallogr F Struct Biol Commun 72: 619-626
- PubMed: 27487926
- DOI: https://doi.org/10.1107/S2053230X16011018
- Primary Citation of Related Structures:
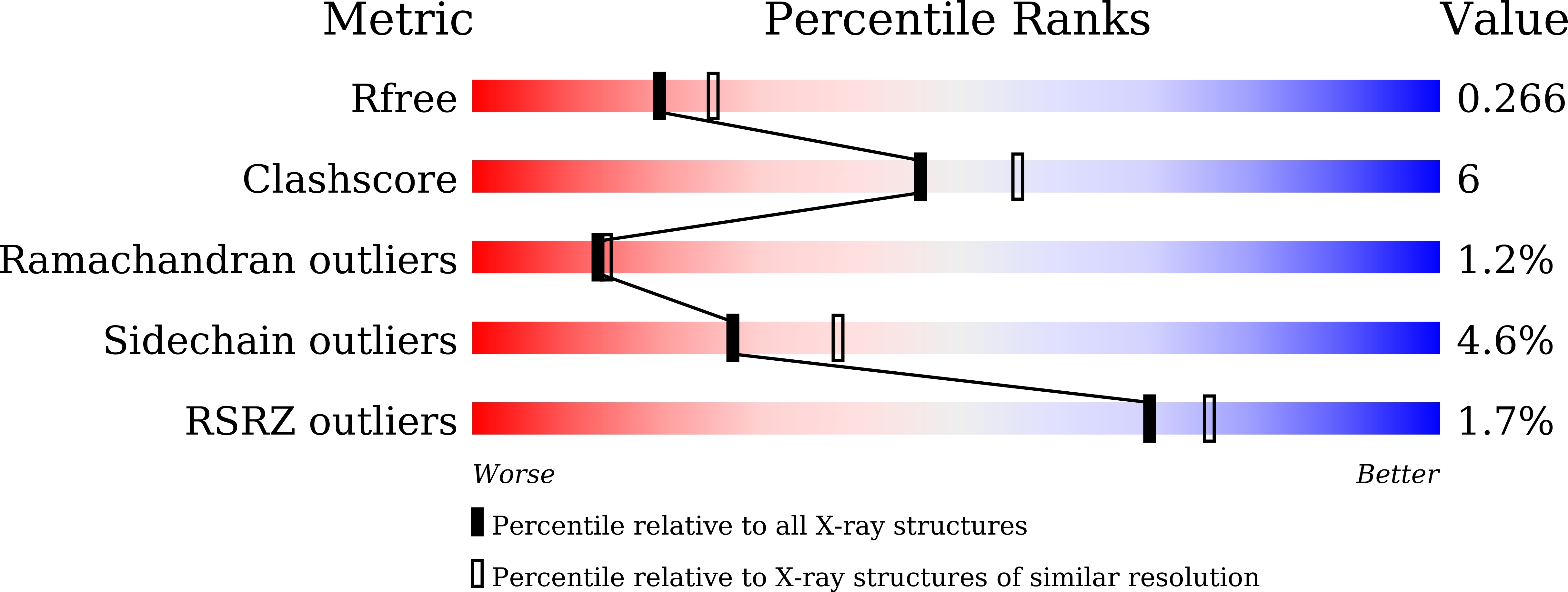
5B4N - PubMed Abstract:
The SCF ubiquitin ligase comprises four components: Skp1, Cul1, Rbx1 and a variable-subunit F-box protein. The F-box protein Fbs1, which recognizes the N-linked glycoproteins, is involved in the endoplasmic reticulum-associated degradation pathway. Although FBG3, another F-box protein, shares 51% sequence identity with Fbs1, FBG3 does not bind glycoproteins. To investigate the sequence-structure relationship of the substrate-binding pocket, the crystal structure of a mutant substrate-binding domain of Fbs1 in which the six nonconserved regions (β1, β2-β3, β3-β4, β5-β6, β7-β8 and β9-β10) of Fbs1 were substituted with those of FBG3 was determined. The substrate-binding pocket of this model exhibits structural features that differ from those of Fsb1.
Organizational Affiliation:
Picobiology Institute, Graduate School of Life Science, University of Hyogo, 3-2-1 Kouto, Kamigori-cho, Ako-gun, Hyogo 678-1297, Japan.