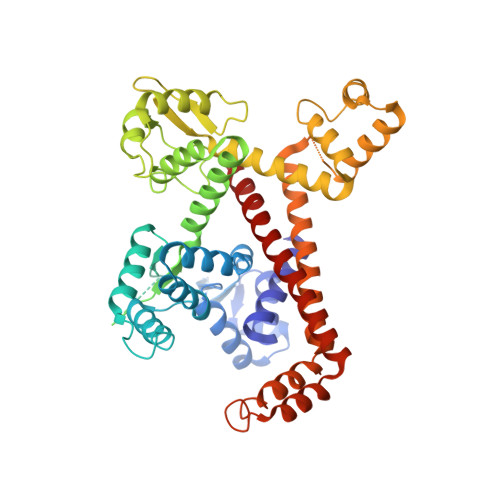
Structural Analysis of Human Rpc32Beta - Rpc62 Complex.
Boissier, F., Dumay-Odelot, H., Teichmann, M., Fribourg, S.(2015) J Struct Biol 192: 313
- PubMed: 26394183
- DOI: https://doi.org/10.1016/j.jsb.2015.09.004
- Primary Citation of Related Structures:
5AFQ - PubMed Abstract:
Transcription initiation by eukaryotic RNA polymerase (Pol) III relies on the subcomplex RPC62/RPC39/RPC32. Two distinct isoforms of RPC32 are encoded in the human genome. RPC32α expression is highly regulated and found only in stem cells and transformed cells, whereas RPC32β is ubiquitously expressed in tissues. Here we identify a core-interacting domain of RPC32 sufficient for the interaction with RPC62. We present the crystal structure of a complex of RPC62 and the RPC32β core domain. RPC32β associates with the extended winged helix 1 and 2 and the coiled coil domain of RPC62 qualifying RPC32 as a molecular bridge in between RPC62 domains. The RPC62-RPC32 complex fit into EM data suggests a bi-functional role for RPC32 through interactions with the largest Pol III subunit and through solvent exposed residues. RPC32 positioning into Pol III suggests that subunit-specific contacts at the surface of the Pol III holoenzyme are critical for its function.
Organizational Affiliation:
Université de Bordeaux, Institut Européen de Chimie et Biologie, ARNA Laboratory, F-33607 Pessac, France; Institut National de la Santé Et de la Recherche Médicale, INSERM - U869, ARNA Laboratory, F-33000 Bordeaux, France.