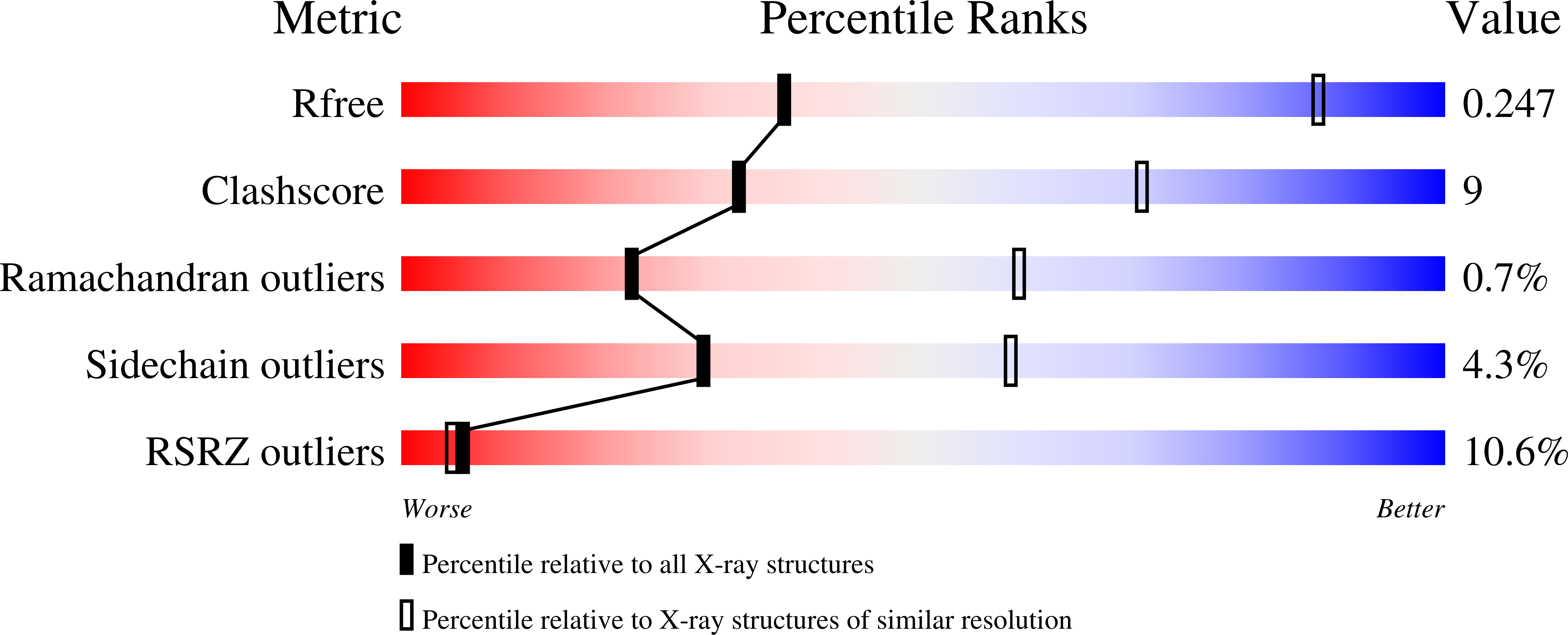
Crystal Structure of the Dynamin Tetramer
Reubold, T.F., Faelber, K., Plattner, N., Posor, Y., Ketel, K., Curth, U., Schlegel, J., Anand, R., Manstein, D.J., Noe, F., Haucke, V., Daumke, O., Eschenburg, S.(2015) Nature 525: 404
- PubMed: 26302298
- DOI: https://doi.org/10.1038/nature14880
- Primary Citation of Related Structures:
5A3F - PubMed Abstract:
The mechanochemical protein dynamin is the prototype of the dynamin superfamily of large GTPases, which shape and remodel membranes in diverse cellular processes. Dynamin forms predominantly tetramers in the cytosol, which oligomerize at the neck of clathrin-coated vesicles to mediate constriction and subsequent scission of the membrane. Previous studies have described the architecture of dynamin dimers, but the molecular determinants for dynamin assembly and its regulation have remained unclear. Here we present the crystal structure of the human dynamin tetramer in the nucleotide-free state. Combining structural data with mutational studies, oligomerization measurements and Markov state models of molecular dynamics simulations, we suggest a mechanism by which oligomerization of dynamin is linked to the release of intramolecular autoinhibitory interactions. We elucidate how mutations that interfere with tetramer formation and autoinhibition can lead to the congenital muscle disorders Charcot-Marie-Tooth neuropathy and centronuclear myopathy, respectively. Notably, the bent shape of the tetramer explains how dynamin assembles into a right-handed helical oligomer of defined diameter, which has direct implications for its function in membrane constriction.
Organizational Affiliation:
Institut für Biophysikalische Chemie, Medizinische Hochschule Hannover, Carl-Neuberg-Str. 1, 30625 Hannover, Germany.