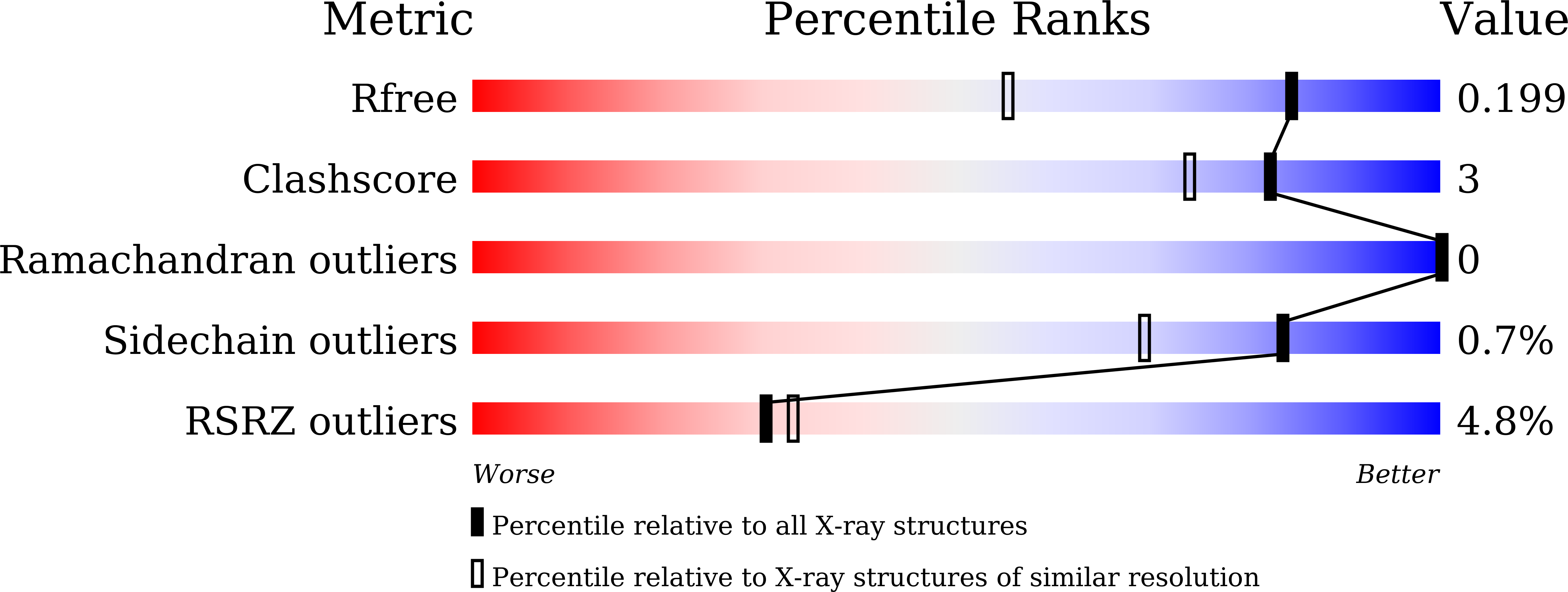
Crystal structures of two forms of the Acanthamoeba polyphaga mimivirus Rab GTPase
Ku, B., You, J.A., Oh, K.J., Yun, H.Y., Lee, H.S., Shin, H.C., Jung, J., Shin, Y.B., Kim, S.J.(2017) Arch Virol 162: 3407-3416
- PubMed: 28779233
- DOI: https://doi.org/10.1007/s00705-017-3510-2
- Primary Citation of Related Structures:
5XC3, 5XC5 - PubMed Abstract:
Acanthamoeba polyphaga mimivirus (APMV) is a member of the family of giant viruses, harboring a 1,200 kbp genome within its 700 nm-diameter viral particle. The R214 gene of the APMV genome was recently shown to encode a homologue of the Rab GTPases, molecular switch proteins known to play a pivotal role in the regulation of membrane trafficking that were considered to exist only in eukaryotes. Herein, we report the first crystal structures of GDP- and GTP-bound forms of APMV Rab GTPase, both of which were determined at high resolution. An in-depth structural comparison of APMV Rab with each other and with mammalian Rab homologues led to an atomic-level elucidation of the inactive-active conformational change upon GDP/GTP exchange. APMV Rab GTPase exhibited considerable structural similarity to human Rab5, as previously predicted based on its amino acid sequence. However, it also contains unique structural features differentiating it from mammalian homologues, such as the functional substitution of a phenylalanine residue for the stabilization of the nucleotide's guanine base.
Organizational Affiliation:
Disease Target Structure Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, 34141, Republic of Korea. bku@kribb.re.kr.