The Structure of an Oxalate Oxidoreductase Provides Insight into Microbial 2-Oxoacid Metabolism.
Gibson, M.I., Brignole, E.J., Pierce, E., Can, M., Ragsdale, S.W., Drennan, C.L.(2015) Biochemistry 54: 4112-4120
- PubMed: 26061898
- DOI: https://doi.org/10.1021/acs.biochem.5b00521
- Primary Citation of Related Structures:
5C4I - PubMed Abstract:
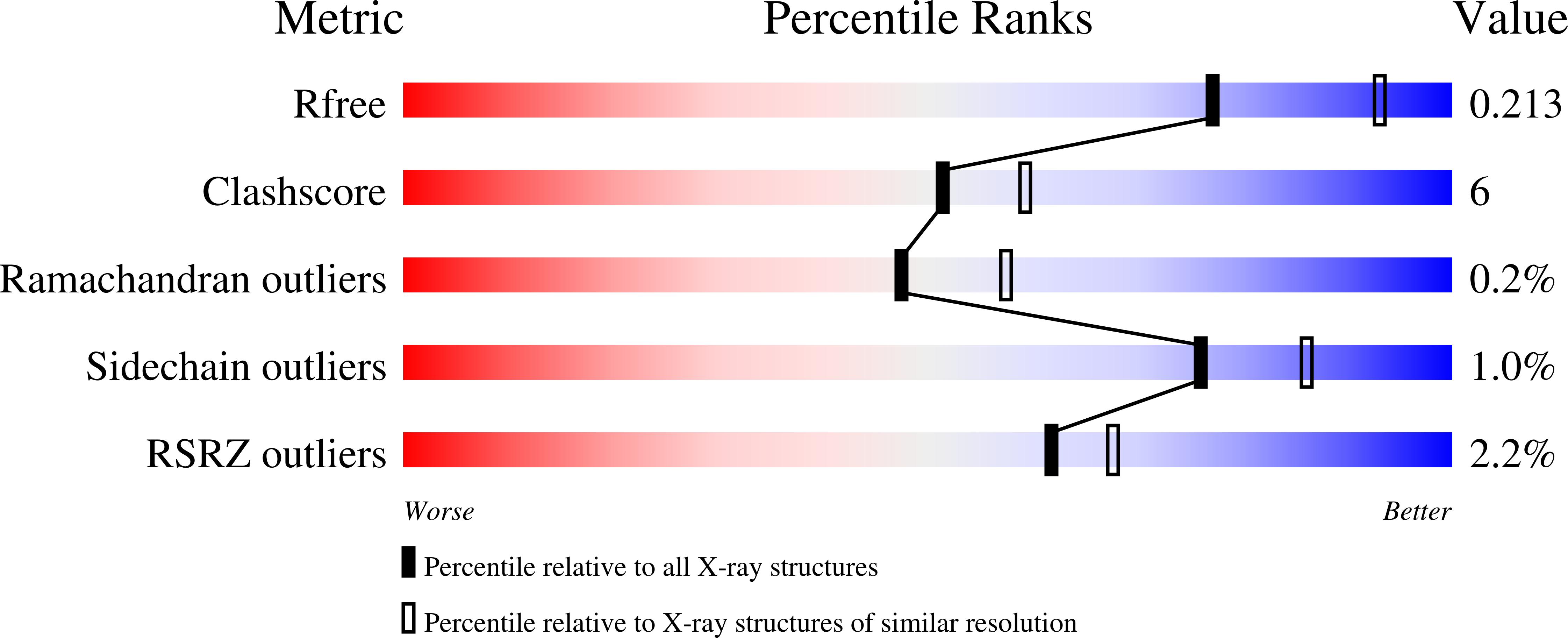
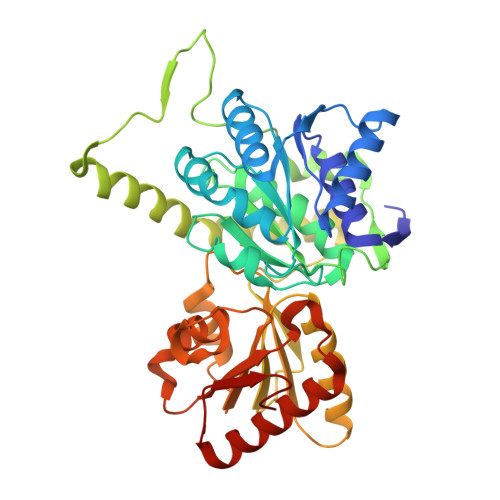
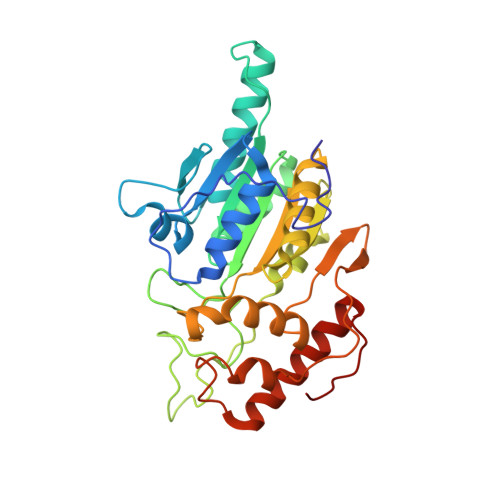
Thiamine pyrophosphate (TPP), a derivative of vitamin B1, is a versatile and ubiquitous cofactor. When coupled with [4Fe-4S] clusters in microbial 2-oxoacid:ferredoxin oxidoreductases (OFORs), TPP is involved in catalyzing low-potential redox reactions that are important for the synthesis of key metabolites and the reduction of N2, H(+), and CO2. We have determined the high-resolution (2.27 Å) crystal structure of the TPP-dependent oxalate oxidoreductase (OOR), an enzyme that allows microbes to grow on oxalate, a widely occurring dicarboxylic acid that is found in soil and freshwater and is responsible for kidney stone disease in humans. OOR catalyzes the anaerobic oxidation of oxalate, harvesting the low-potential electrons for use in anaerobic reduction and fixation of CO2. We compare the OOR structure to that of the only other structurally characterized OFOR family member, pyruvate:ferredoxin oxidoreductase. This side-by-side structural analysis highlights the key similarities and differences that are relevant for the chemistry of this entire class of TPP-utilizing enzymes.
Organizational Affiliation:
†Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139, United States.