Structure-based design of a bisphosphonate 5'(3')-deoxyribonucleotidase inhibitor
Pachl, P., Simak, O., Rezacova, P., Fabry, M., Budesinsky, M., Rosenberg, I., Brynda, J.(2015) Medchemcomm 6: 1635-1638
Experimental Data Snapshot
(2015) Medchemcomm 6: 1635-1638
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'(3')-deoxyribonucleotidase, cytosolic type | 195 | Homo sapiens | Mutation(s): 0 Gene Names: NT5C, DNT1, UMPH2 EC: 3.1.3 | 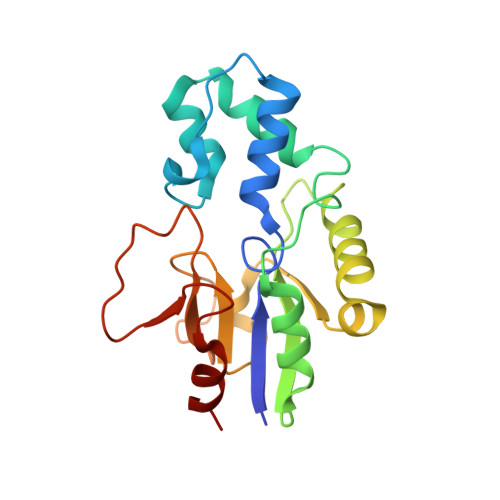 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8TCD5 (Homo sapiens) Explore Q8TCD5 Go to UniProtKB: Q8TCD5 | |||||
PHAROS: Q8TCD5 GTEx: ENSG00000125458 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8TCD5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 2O2 Query on 2O2 | E [auth A], I [auth B] | 1-{2-deoxy-3,5-O-[phenyl(phosphono)methylidene]-beta-D-threo-pentofuranosyl}-5-[(E)-2-phosphonoethenyl]pyrimidine-2,4(1H,3H)-dione C18 H20 N2 O11 P2 AYSYVLQGVXZPIY-OAIWFRFLSA-N |  | ||
| PO4 Query on PO4 | D [auth A], H [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| GOL Query on GOL | F [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| MG Query on MG | C [auth A], G [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 38.818 | α = 111.18 |
| b = 46.451 | β = 88.16 |
| c = 61.738 | γ = 104.7 |
| Software Name | Purpose |
|---|---|
| MOLREP | phasing |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| XSCALE | data scaling |
| REFMAC | refinement |
| XDS | data scaling |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Grant Agency of the Czech Republic | Czech Republic | 15-05677S |