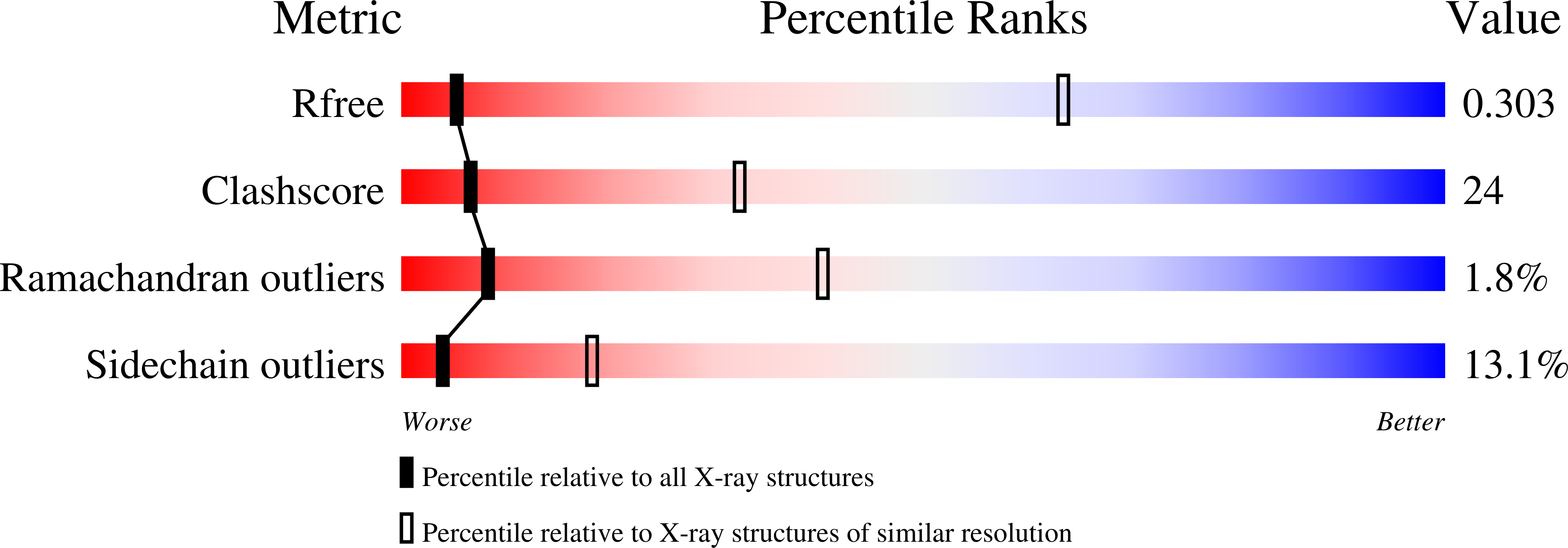Structure of an 'open' clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport.
Laponogov, I., Veselkov, D.A., Crevel, I.M., Pan, X.S., Fisher, L.M., Sanderson, M.R.(2013) Nucleic Acids Res 41: 9911-9923
- PubMed: 23965305
- DOI: https://doi.org/10.1093/nar/gkt749
- Primary Citation of Related Structures:
4I3H, 4JUO - PubMed Abstract:
Type II topoisomerases regulate DNA supercoiling and chromosome segregation. They act as ATP-operated clamps that capture a DNA duplex and pass it through a transient DNA break in a second DNA segment via the sequential opening and closure of ATPase-, G-DNA- and C-gates. Here, we present the first 'open clamp' structures of a 3-gate topoisomerase II-DNA complex, the seminal complex engaged in DNA recognition and capture. A high-resolution structure was solved for a (full-length ParE-ParC55)2 dimer of Streptococcus pneumoniae topoisomerase IV bound to two DNA molecules: a closed DNA gate in a B-A-B form double-helical conformation and a second B-form duplex associated with closed C-gate helices at a novel site neighbouring the catalytically important β-pinwheel DNA-binding domain. The protein N gate is present in an 'arms-wide-open' state with the undimerized N-terminal ParE ATPase domains connected to TOPRIM domains via a flexible joint and folded back allowing ready access both for gate and transported DNA segments and cleavage-stabilizing antibacterial drugs. The structure shows the molecular conformations of all three gates at 3.7 Å, the highest resolution achieved for the full complex to date, and illuminates the mechanism of DNA capture and transport by a type II topoisomerase.
Organizational Affiliation:
Randall Division of Cell and Molecular Biophysics, 3rd floor New Hunt's House, Guy's Campus, King's College London, London, SE1 1UL, UK and Division of Biomedical Sciences, St. George's, University of London, Cranmer Terrace, London, SW17 0RE, UK.