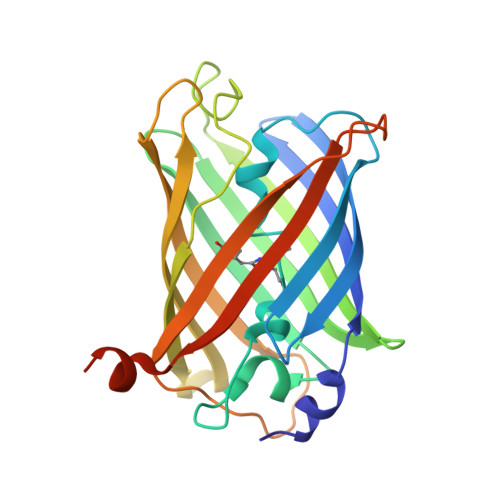
Yellow fluorescent protein phiYFPv (Phialidium): structure and structure-based mutagenesis.
Pletneva, N.V., Pletnev, V.Z., Souslova, E., Chudakov, D.M., Lukyanov, S., Martynov, V.I., Arhipova, S., Artemyev, I., Wlodawer, A., Dauter, Z., Pletnev, S.(2013) Acta Crystallogr D Biol Crystallogr 69: 1005-1012
- PubMed: 23695245
- DOI: https://doi.org/10.1107/S0907444913004034
- Primary Citation of Related Structures:
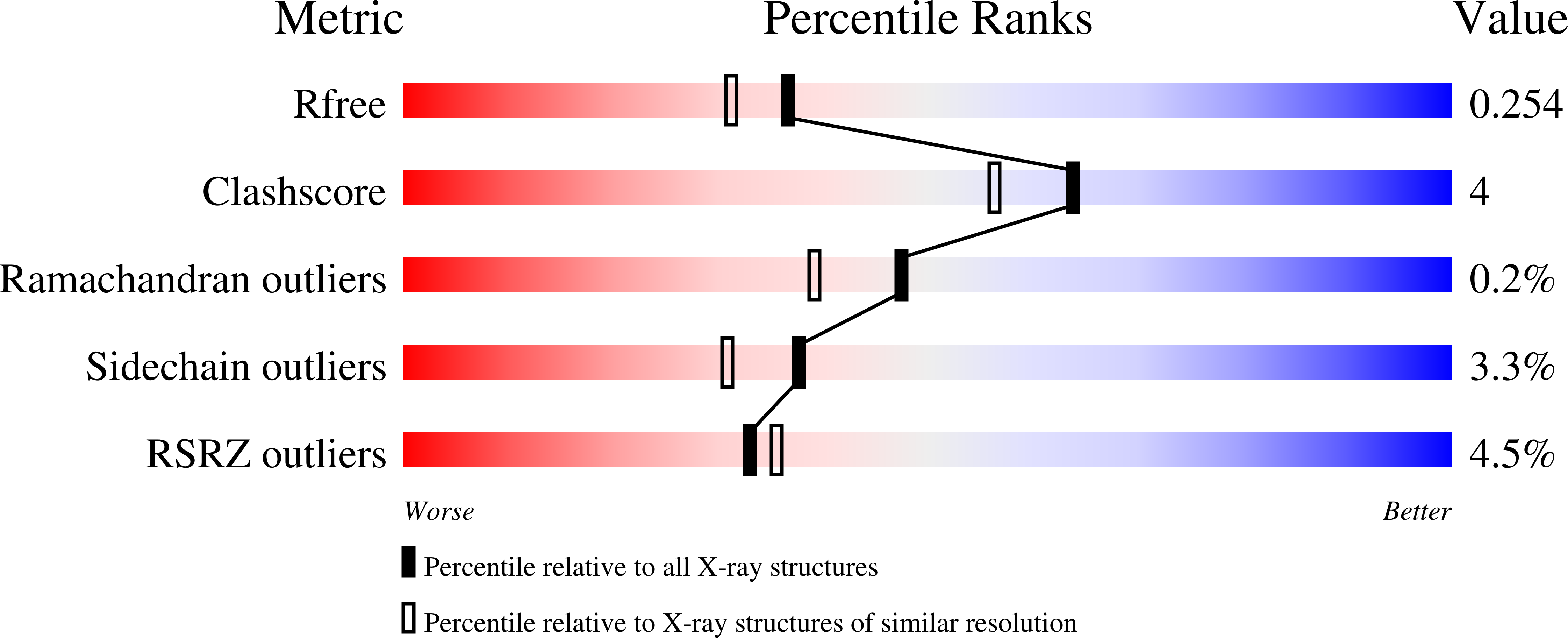
4HE4 - PubMed Abstract:
The yellow fluorescent protein phiYFPv (λem(max) ≃ 537 nm) with improved folding has been developed from the spectrally identical wild-type phiYFP found in the marine jellyfish Phialidium. The latter fluorescent protein is one of only two known cases of naturally occurring proteins that exhibit emission spectra in the yellow-orange range (535-555 nm). Here, the crystal structure of phiYFPv has been determined at 2.05 Å resolution. The `yellow' chromophore formed from the sequence triad Thr65-Tyr66-Gly67 adopts the bicyclic structure typical of fluorophores emitting in the green spectral range. It was demonstrated that perfect antiparallel π-stacking of chromophore Tyr66 and the proximal Tyr203, as well as Val205, facing the chromophore phenolic ring are chiefly responsible for the observed yellow emission of phiYFPv at 537 nm. Structure-based site-directed mutagenesis has been used to identify the key functional residues in the chromophore environment. The obtained results have been utilized to improve the properties of phiYFPv and its homologous monomeric biomarker tagYFP.
Organizational Affiliation:
Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russian Federation.