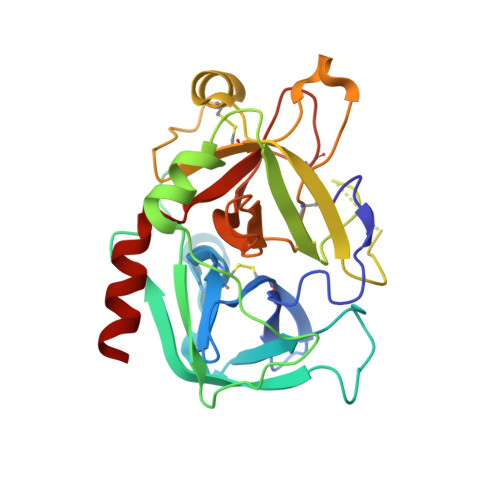
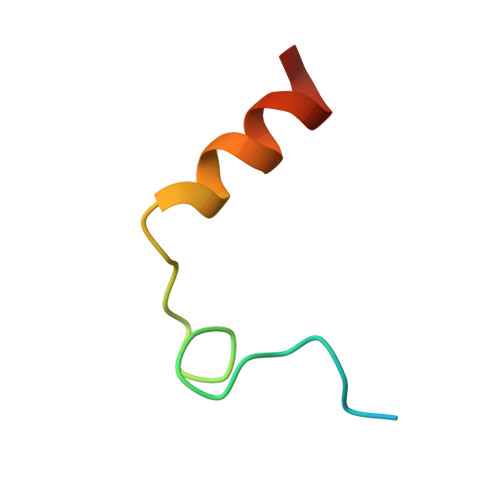
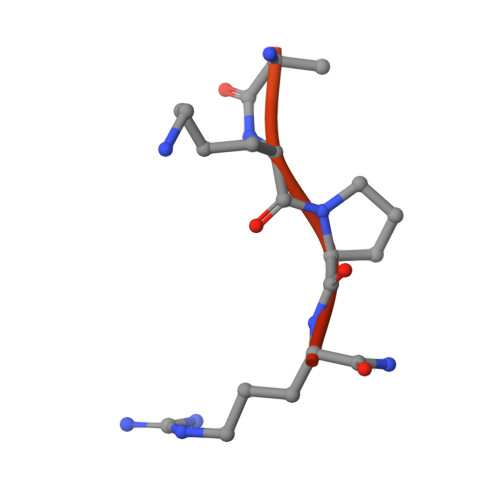
The Tick-Derived Anticoagulant Madanin is Processed by Thrombin and Factor Xa.
Figueiredo, A.C., De Sanctis, D., Pereira, P.J.(2013) PLoS One 8: 71866
- PubMed: 23951260
- DOI: https://doi.org/10.1371/journal.pone.0071866
- Primary Citation of Related Structures:
4BOH - PubMed Abstract:
The cysteine-less peptidic anticoagulants madanin-1 and madanin-2 from the bush tick Haemaphysalis longicornis are the founding members of the MEROPS inhibitor family I53. It has been previously suggested that madanins exert their functional activity by competing with physiological substrates for binding to the positively charged exosite I (fibrinogen-binding exosite) of α-thrombin. We hereby demonstrate that competitive inhibition of α-thrombin by madanin-1 or madanin-2 involves binding to the enzyme's active site. Moreover, the blood coagulation factors IIa and Xa are shown to hydrolyze both inhibitors at different, although partially overlapping cleavage sites. Finally, the three-dimensional structure of the complex formed between human α-thrombin and a proteolytic fragment of madanin-1, determined by X-ray crystallography, elucidates the molecular details of madanin-1 recognition and processing by the proteinase. Taken together, the current findings establish the mechanism of action of madanins, natural anticoagulants that behave as cleavable competitive inhibitors of thrombin.
Organizational Affiliation:
IBMC - Instituto de Biologia Molecular e Celular, Universidade do Porto, Porto, Portugal.