Liesegang-Like Patterns of Toll Crystals Grown in Gel.
Gangloff, M., Moreno, A., Gay, N.J.(2013) J Appl Crystallogr 46: 337
- PubMed: 23596340
- DOI: https://doi.org/10.1107/S0021889812051606
- Primary Citation of Related Structures:
4ARR - PubMed Abstract:
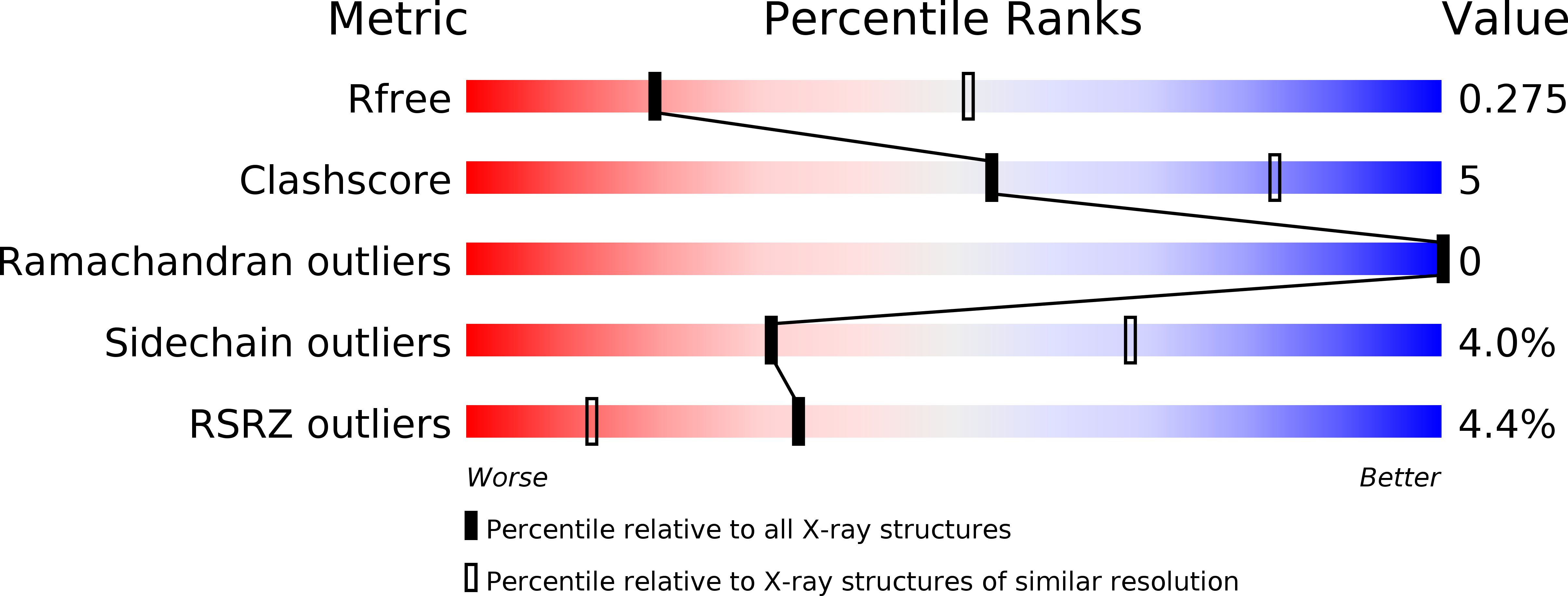
Generating high-quality crystals remains a bottleneck in biological and materials sciences. Here a counter-diffusion method was used to improve the X-ray diffraction quality of the N-terminal domain of Drosophila melanogaster Toll receptor crystals. It was observed that crystallization occurred with a peculiar pattern along the capillary resembling Liesegang bands; this phenomenon is described at both macroscopic and atomic levels. It was found that bands appeared for native protein as well as for co-crystals of magic triangle (I3C)-bound protein even though they crystallize in different space groups. Crystallization occurred with a linear recurrence independent of the precipitant concentration and a protein-specific spacing coefficient. Bandwidth varied along the capillary, oscillating between large precipitation areas and single crystals. The reported data suggest that repetitive patterns can be generated with biological macromolecules in the presence of sodium malonate as a crystallization agent. A comparison with typical Liesegang patterns and the possible mechanism underlying this phenomenon are discussed.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, Cambridge CB2 1GA, UK.