Crystal structure of Mycobacterium tuberculosis ketol-acid reductoisomerase at 1.0 angstrom resolution - a potential target for anti-tuberculosis drug discovery.
Lv, Y., Kandale, A., Wun, S.J., McGeary, R.P., Williams, S.J., Kobe, B., Sieber, V., Schembri, M.A., Schenk, G., Guddat, L.W.(2016) FEBS J 283: 1184-1196
- PubMed: 26876563
- DOI: https://doi.org/10.1111/febs.13672
- Primary Citation of Related Structures:
4YPO - PubMed Abstract:
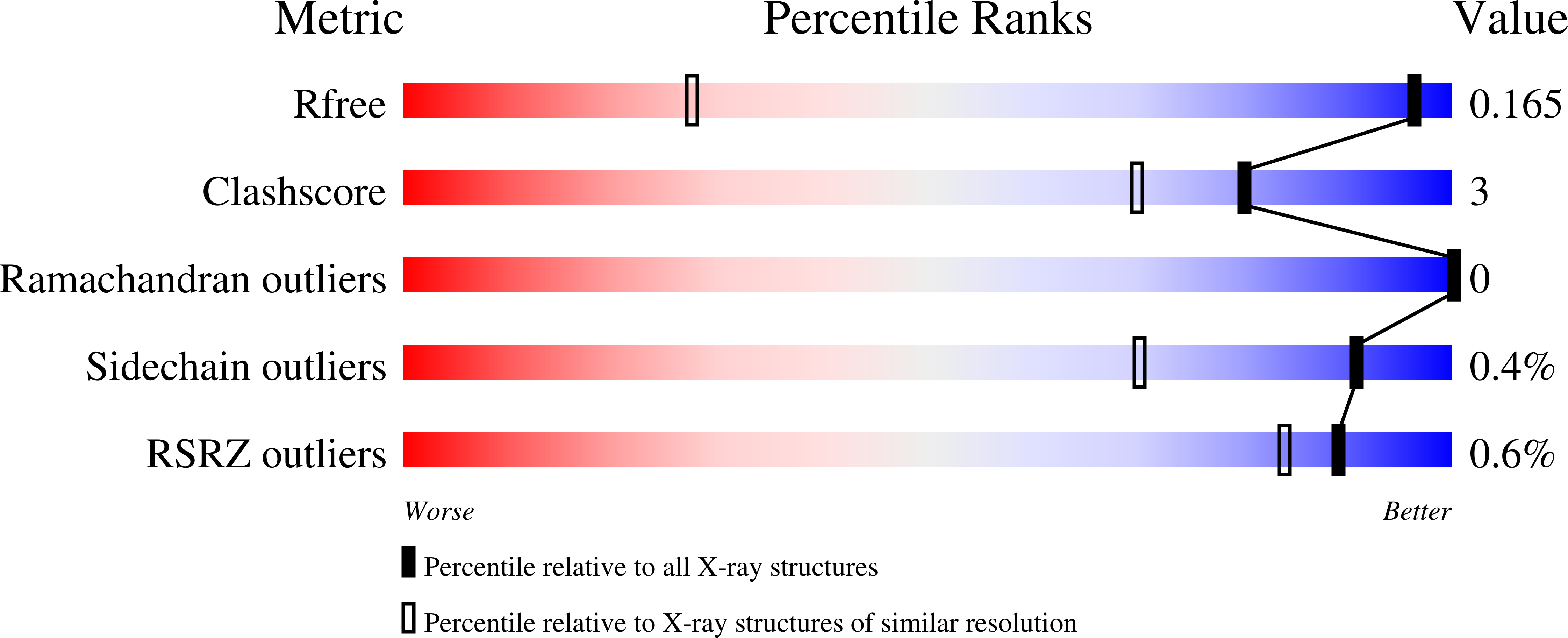
The biosynthetic pathway for the branched-chain amino acids is present in plants, fungi and bacteria, but not in animals, making it an attractive target for herbicidal and antimicrobial drug discovery. Ketol-acid reductoisomerase (KARI; EC 1.1.1.86) is the second enzyme in this pathway, converting in a Mg(2+) - and NADPH-dependent reaction either 2-acetolactate or 2-aceto-2-hydroxybutyrate to their corresponding 2,3-dihydroxy-3-alkylbutyrate products. Here, we have determined the crystal structure of Mycobacterium tuberculosis (Mt) KARI, a class I KARI, with two magnesium ions bound in the active site. X-ray data were obtained to 1.0 Å resolution and the final model has an Rfree of 0.163. The structure shows that the active site is solvent-accessible with the two metal ions separated by 4.7 Å. A comparison of this structure with that of Mg(2+) -free Pseudomonas aeruginosa KARI suggests that upon magnesium binding no movement of the N domain relative to the C domain occurs. However, upon formation of the Michaelis complex, as illustrated in the structure of Slackia exigua KARI in complex with NADH.Mg(2+) . N-hydroxy-N-isopropyloxamate (IpOHA, a transition state analog), domain movements and reduction of the metal-metal distance to 3.5 Å are observed. This inherent flexibility therefore appears to be critical for initiation of the KARI-catalyzed reaction. This study provides new insights into the complex structural rearrangements required for activity of KARIs, particularly those belonging to class I, and provides the framework for the rational design of Mt KARI inhibitors that can be tested as novel antituberculosis agents. Coordinates and structure factors for the Mt KARI.Mg(2+) complex are available in the Protein Data Bank under accession number 4YPO.
Organizational Affiliation:
School of Chemistry and Molecular Biosciences and Australian Infectious Disease Research Centre, University of Queensland, Brisbane, Australia.