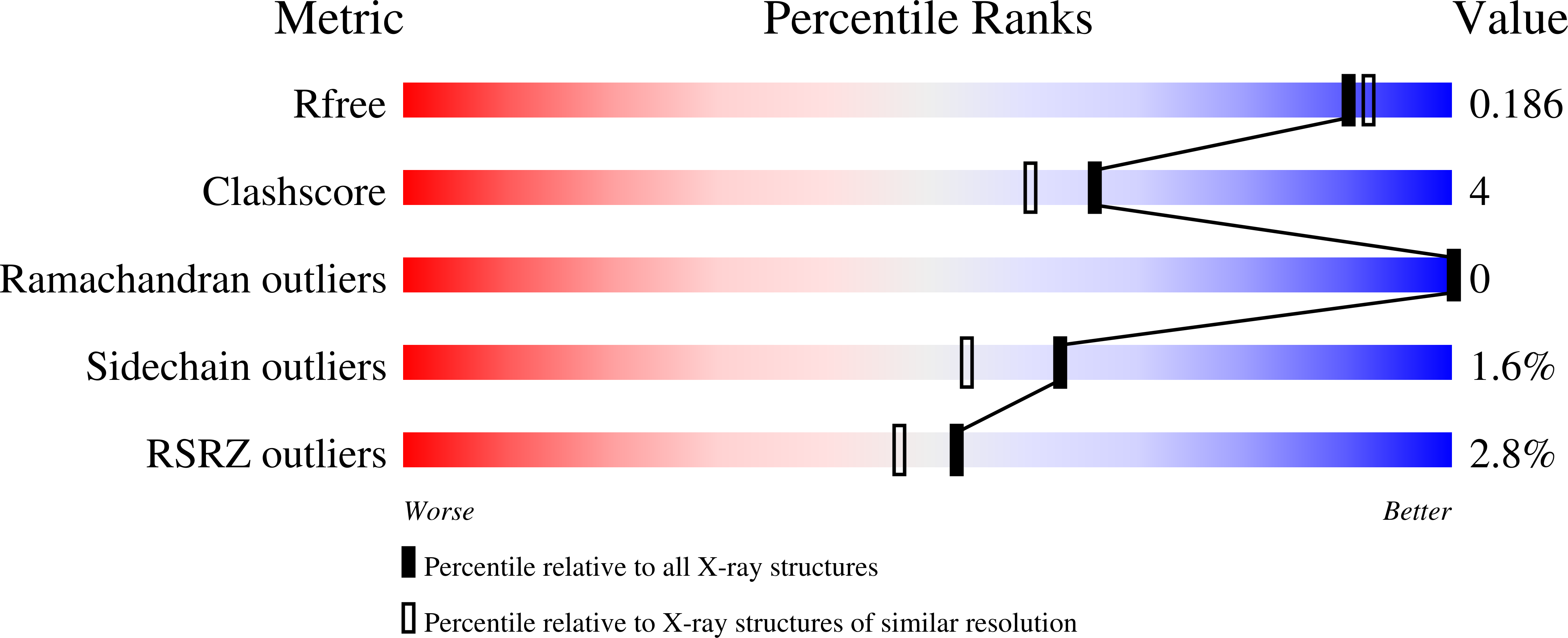
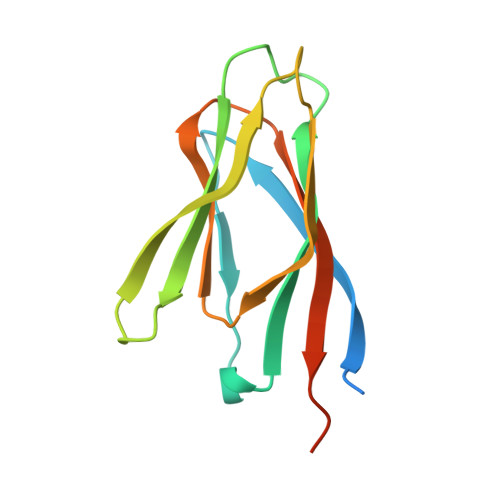
Structural polymorphism in the N-terminal oligomerization domain of NPM1.
Mitrea, D.M., Grace, C.R., Buljan, M., Yun, M.K., Pytel, N.J., Satumba, J., Nourse, A., Park, C.G., Madan Babu, M., White, S.W., Kriwacki, R.W.(2014) Proc Natl Acad Sci U S A 111: 4466-4471
- PubMed: 24616519
- DOI: https://doi.org/10.1073/pnas.1321007111
- Primary Citation of Related Structures:
4N8M - PubMed Abstract:
Nucleophosmin (NPM1) is a multifunctional phospho-protein with critical roles in ribosome biogenesis, tumor suppression, and nucleolar stress response. Here we show that the N-terminal oligomerization domain of NPM1 (Npm-N) exhibits structural polymorphism by populating conformational states ranging from a highly ordered, folded pentamer to a highly disordered monomer. The monomer-pentamer equilibrium is modulated by posttranslational modification and protein binding. Phosphorylation drives the equilibrium in favor of monomeric forms, and this effect can be reversed by Npm-N binding to its interaction partners. We have identified a short, arginine-rich linear motif in NPM1 binding partners that mediates Npm-N oligomerization. We propose that the diverse functional repertoire associated with NPM1 is controlled through a regulated unfolding mechanism signaled through posttranslational modifications and intermolecular interactions.
Organizational Affiliation:
Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN 38105.