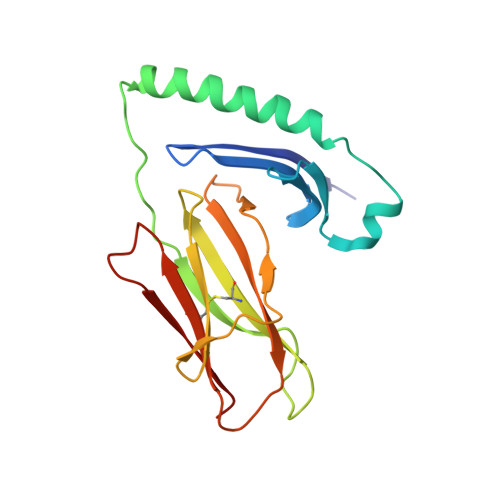
Structure-Based Design of Altered MHC Class II-Restricted Peptide Ligands with Heterogeneous Immunogenicity.
Chen, S., Li, Y., Depontieu, F.R., McMiller, T.L., English, A.M., Shabanowitz, J., Kos, F., Sidney, J., Sette, A., Rosenberg, S.A., Hunt, D.F., Mariuzza, R.A., Topalian, S.L.(2013) J Immunol 191: 5097-5106
- PubMed: 24108701
- DOI: https://doi.org/10.4049/jimmunol.1300467
- Primary Citation of Related Structures:
4IS6 - PubMed Abstract:
Insights gained from characterizing MHC-peptide-TCR interactions have held the promise that directed structural modifications can have predictable functional consequences. The ability to manipulate T cell reactivity synthetically or through genetic engineering might thus be translated into new therapies for common diseases such as cancer and autoimmune disorders. In the current study, we determined the crystal structure of HLA-DR4 in complex with the nonmutated dominant gp100 epitope gp10044-59, associated with many melanomas. Altered peptide ligands (APLs) were designed to enhance MHC binding and hence T cell recognition of gp100 in HLA-DR4(+) melanoma patients. Increased MHC binding of several APLs was observed, validating this approach biochemically. Nevertheless, heterogeneous preferences of CD4(+) T cells from several HLA-DR4(+) melanoma patients for different gp100 APLs suggested highly variable TCR usage, even among six patients who had been vaccinated against the wild-type gp100 peptide. This heterogeneity prevented the selection of an APL candidate for developing an improved generic gp100 vaccine in melanoma. Our results are consistent with the idea that even conservative changes in MHC anchor residues may result in subtle, yet crucial, effects on peptide contacts with the TCR or on peptide dynamics, such that alterations intended to enhance immunogenicity may be unpredictable or counterproductive. They also underscore a critical knowledge gap that needs to be filled before structural and in vitro observations can be used reliably to devise new immunotherapies for cancer and other disorders.
Organizational Affiliation:
Department of Surgery, Johns Hopkins University School of Medicine and the Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD 21287;