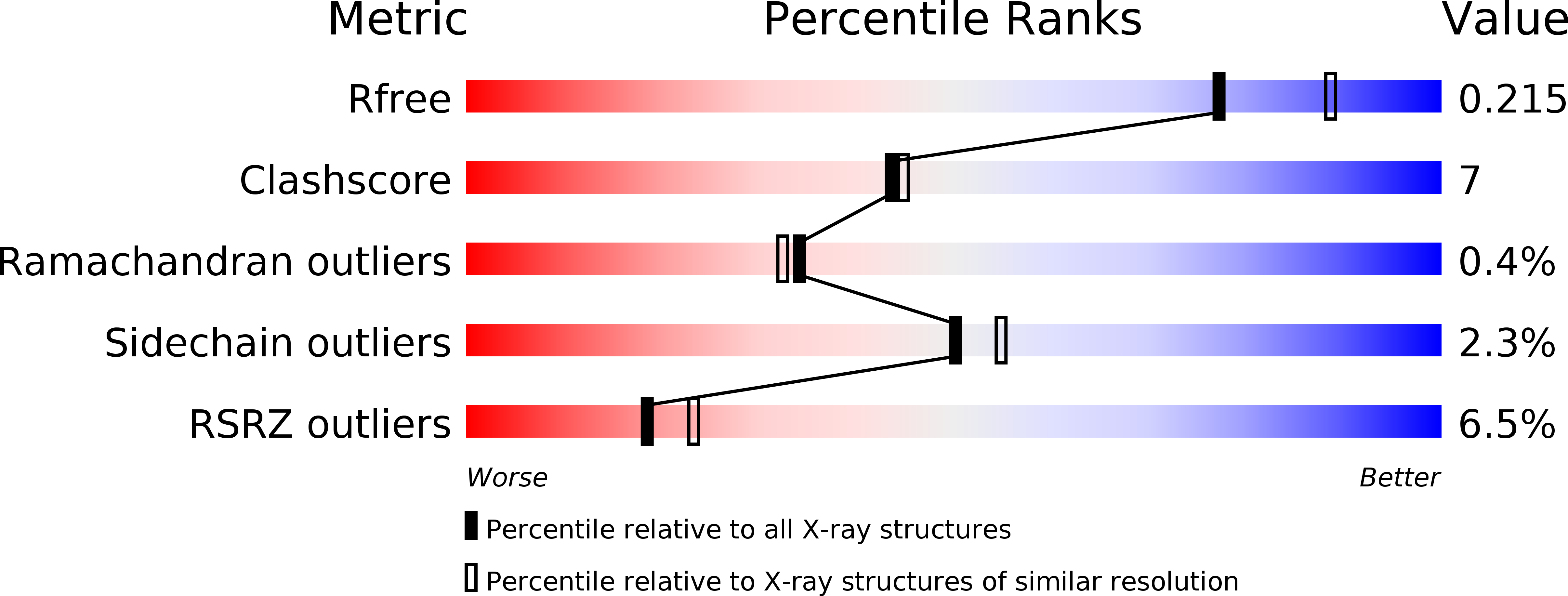
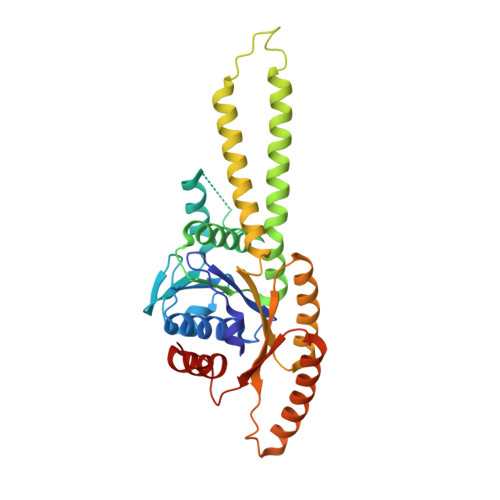
A Conserved Archaeal Pathway for Tail-Anchored Membrane Protein Insertion.
Sherrill, J., Mariappan, M., Dominik, P., Hegde, R.S., Keenan, R.J.(2011) Traffic 12: 1119
- PubMed: 21658170
- DOI: https://doi.org/10.1111/j.1600-0854.2011.01229.x
- Primary Citation of Related Structures:
3ZQ6 - PubMed Abstract:
Eukaryotic tail-anchored (TA) membrane proteins are inserted into the endoplasmic reticulum by a post-translational TRC40 pathway, but no comparable pathway is known in other domains of life. The crystal structure of an archaebacterial TRC40 sequence homolog bound to ADP•AlF(4) (-) reveals characteristic features of eukaryotic TRC40, including a zinc-mediated dimer and a large hydrophobic groove. Moreover, archaeal TRC40 interacts with the transmembrane domain of TA substrates and directs their membrane insertion. Thus, the TRC40 pathway is more broadly conserved than previously recognized.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology, University of Chicago, Gordon Center for Integrative Science, Room W238, Chicago, IL 60637, USA.