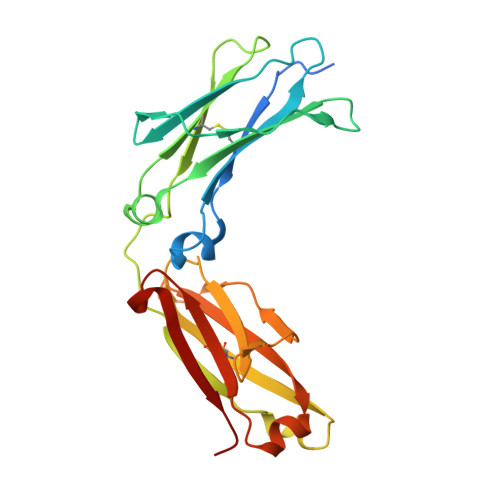
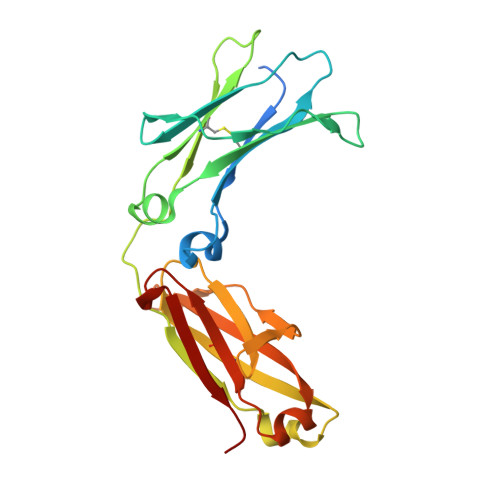
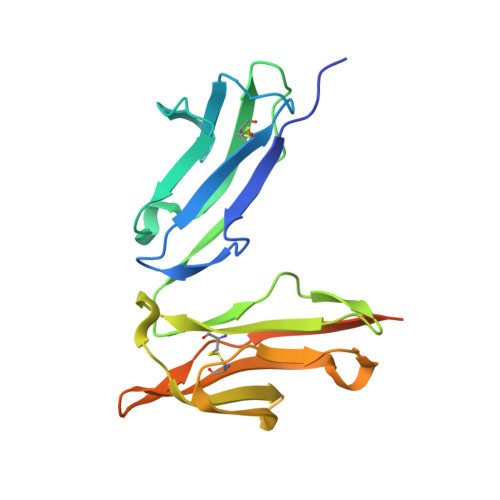
Crystal structure of a novel asymmetrically engineered Fc variant with improved affinity for Fc gamma Rs.
Mimoto, F., Kadono, S., Katada, H., Igawa, T., Kamikawa, T., Hattori, K.(2014) Mol Immunol 58: 132-138
- PubMed: 24334029
- DOI: https://doi.org/10.1016/j.molimm.2013.11.017
- Primary Citation of Related Structures:
3WN5 - PubMed Abstract:
Enhancing the effector function by optimizing the interaction between Fc and Fcγ receptor (FcγR) is a promising approach to enhance the potency of anticancer monoclonal antibodies (mAbs). To date, a variety of Fc engineering approaches to modulate the interaction have been reported, such as afucosylation in the heavy chain Fc region or symmetrically introducing amino acid substitutions into the region, and there is still room to improve FcγR binding and thermal stability of the CH2 domain with these approaches. Recently, we have reported that asymmetric Fc engineering, which introduces different substitutions into each Fc region of heavy chain, can further improve the FcγR binding while maintaining the thermal stability of the CH2 domain by fine-tuning the asymmetric interface between the Fc domain and FcγR. However, the structural mechanism by which the asymmetrically engineered Fc improved FcγR binding remained unclear. In order to elucidate the mechanism, we solved the crystal structure of a novel asymmetrically engineered Fc, asym-mAb23, in complex with FcγRIIIa. Asym-mAb23 has enhanced binding affinity for both FcγRIIIa and FcγRIIa at the highest level of previously reported Fc variants. The structural analysis reveals the features of the asymmetrically engineered Fc in comparison with symmetric Fc and how each asymmetrically introduced substitution contributes to the improved interaction between asym-mAb23 and FcγRIIIa. This crystal structure could be utilized to enable us to design a more potent asymmetric Fc.
Organizational Affiliation:
Research Division, Chugai Pharmaceutical Co., Ltd., Japan.