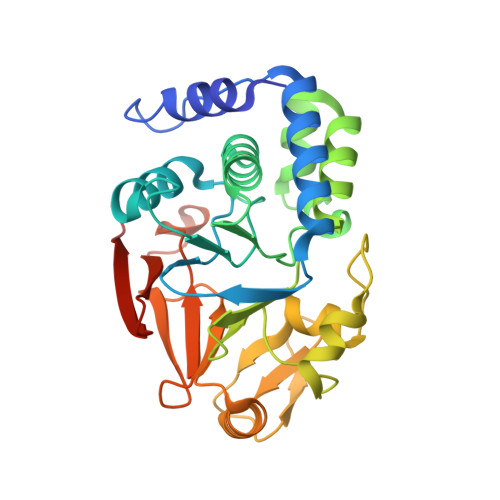
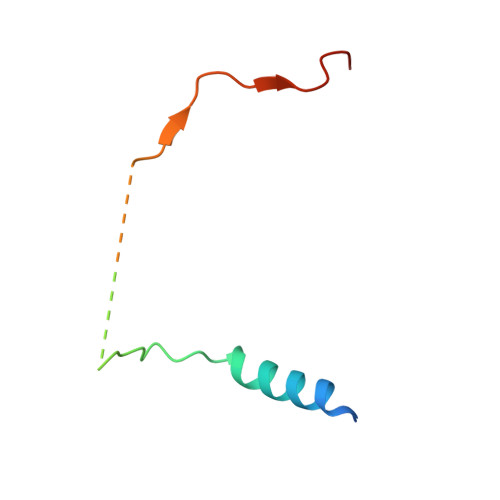
The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme.
O'Connell, N., Nichols, S.R., Heroes, E., Beullens, M., Bollen, M., Peti, W., Page, R.(2012) Structure 20: 1746-1756
- PubMed: 22940584
- DOI: https://doi.org/10.1016/j.str.2012.08.003
- Primary Citation of Related Structures:
3V4Y - PubMed Abstract:
Regulation of protein phosphatase 1 (PP1) is controlled by a diverse array of regulatory proteins. However, how these proteins direct PP1 specificity is not well understood. More than one-third of the nuclear pool of PP1 forms a holoenzyme with the nuclear inhibitor of PP1, NIPP1, to regulate chromatin remodeling, among other essential biological functions. Here, we show that the PP1-binding domain of NIPP1 is an intrinsically disordered protein, which binds PP1 in an unexpected manner. NIPP1 forms an α helix that engages PP1 at a unique interaction site, using polar rather than hydrophobic contacts. Importantly, the structure also reveals a shared PP1 interaction site outside of the RVxF motif, the ΦΦ motif. Finally, we show that NIPP1:PP1 substrate selectivity is determined by altered electrostatics and enhanced substrate localization. Together, our results provide the molecular basis by which NIPP1 directs PP1 substrate specificity in the nucleus.
Organizational Affiliation:
Department of Molecular Pharmacology, Physiology and Biotechnology, Brown University, Providence, RI 02912, USA.