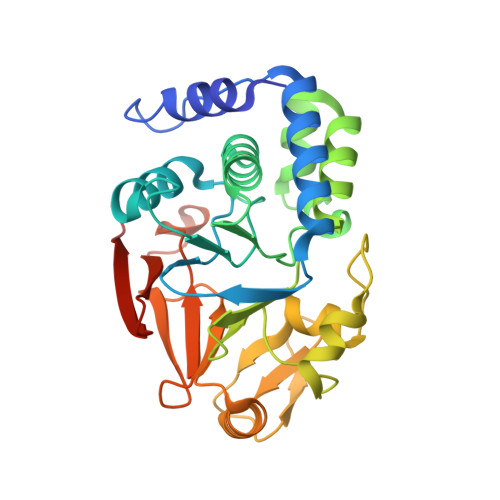
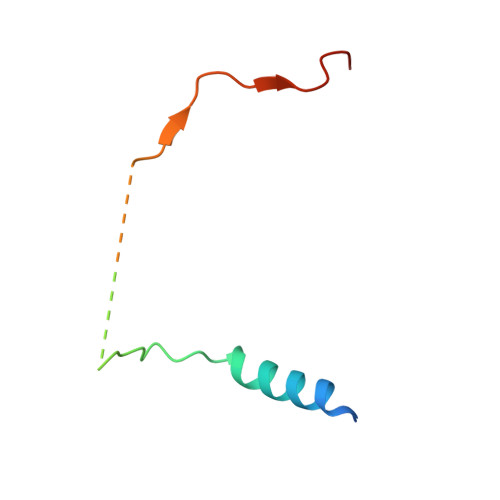
The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme.
O'Connell, N., Nichols, S.R., Heroes, E., Beullens, M., Bollen, M., Peti, W., Page, R.(2012) Structure 20: 1746-1756
- PubMed: 22940584
- DOI: https://doi.org/10.1016/j.str.2012.08.003
- Primary Citation of Related Structures:
3V4Y - PubMed Abstract:
Regulation of protein phosphatase 1 (PP1) is controlled by a diverse array of regulatory proteins. However, how these proteins direct PP1 specificity is not well understood. More than one-third of the nuclear pool of PP1 forms a holoenzyme with the nuclear inhibitor of PP1, NIPP1, to regulate chromatin remodeling, among other essential biological functions. Here, we show that the PP1-binding domain of NIPP1 is an intrinsically disordered protein, which binds PP1 in an unexpected manner. NIPP1 forms an α helix that engages PP1 at a unique interaction site, using polar rather than hydrophobic contacts. Importantly, the structure also reveals a shared PP1 interaction site outside of the RVxF motif, the ΦΦ motif. Finally, we show that NIPP1:PP1 substrate selectivity is determined by altered electrostatics and enhanced substrate localization. Together, our results provide the molecular basis by which NIPP1 directs PP1 substrate specificity in the nucleus.
- Department of Molecular Pharmacology, Physiology and Biotechnology, Brown University, Providence, RI 02912, USA.
Organizational Affiliation: