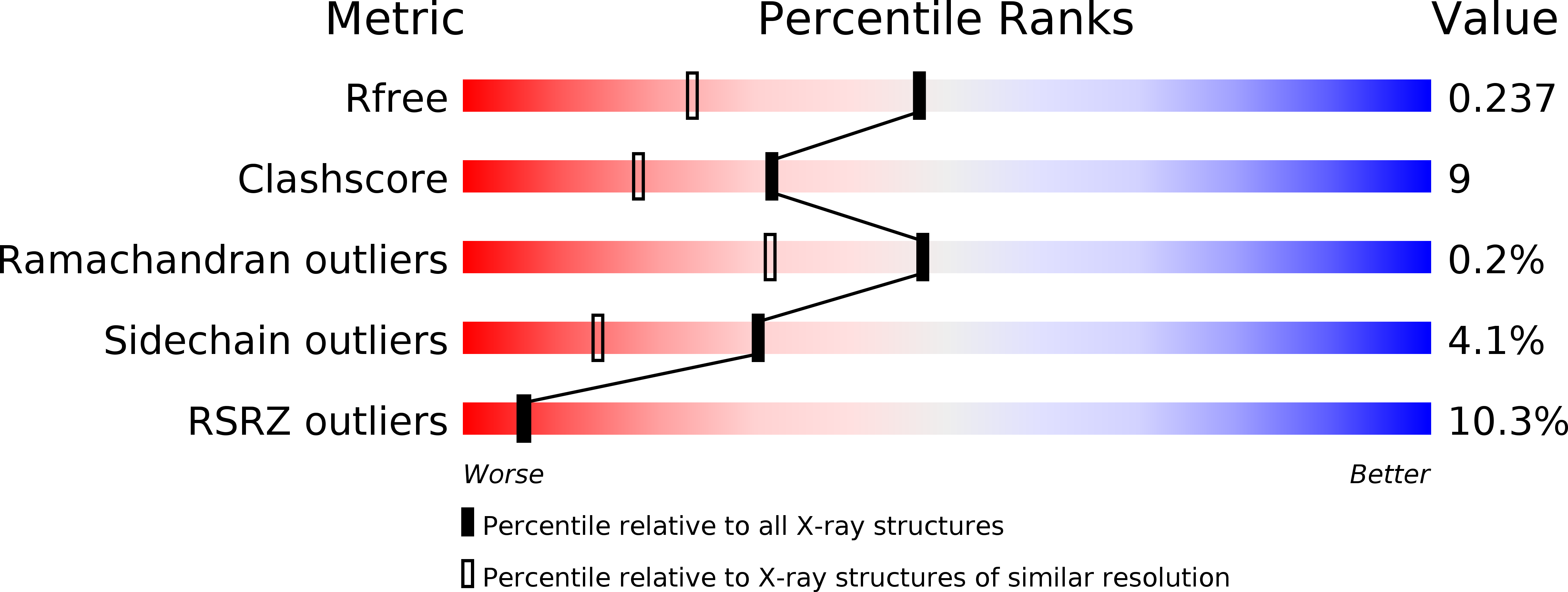
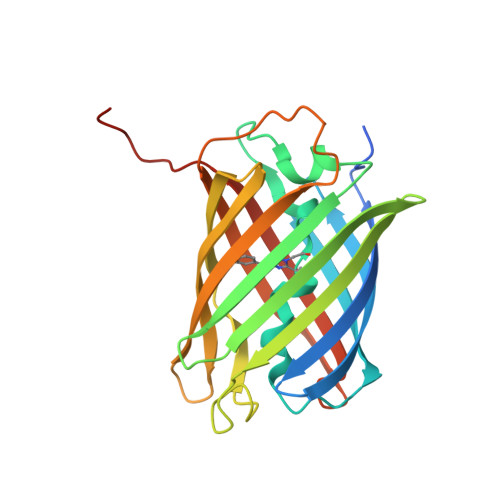
A structural basis for reversible photoswitching of absorbance spectra in red fluorescent protein rsTagRFP.
Pletnev, S., Subach, F.V., Dauter, Z., Wlodawer, A., Verkhusha, V.V.(2012) J Mol Biol 417: 144-151
- PubMed: 22310052
- DOI: https://doi.org/10.1016/j.jmb.2012.01.044
- Primary Citation of Related Structures:
3U8A, 3U8C - PubMed Abstract:
rsTagRFP is the first monomeric red fluorescent protein (FP) with reversibly photoswitchable absorbance spectra. The switching is realized by irradiation of rsTagRFP with blue (440 nm) and yellow (567 nm) light, turning the protein fluorescence ON and OFF, respectively. It is perhaps the most useful probe in this color class that has yet been reported. Because of the photoswitchable absorbance, rsTagRFP can be used as an acceptor in photochromic Förster resonance energy transfer. Yellow FPs, YPet and mVenus, are demonstrated to be excellent photochromic Förster resonance energy transfer donors for the rsTagRFP acceptor in its fusion constructs. Analysis of X-ray structures has shown that photoswitching of rsTagRFP is accompanied by cis-trans isomerization and protonation/deprotonation of the chromophore, with the deprotonated cis- and protonated trans-isomers corresponding to its ON and OFF states, respectively. Unlike in other photoswitchable FPs, both conformers of rsTagRFP chromophore are essentially coplanar. Two other peculiarities of the rsTagRFP chromophore are an essentially hydrophobic environment of its p-hydroxyphenyl site and the absence of direct hydrogen bonding between this moiety and the protein scaffold. The influence of the immediate environment on rsTagRFP chromophore was probed by site-directed mutagenesis. Residues Glu145 and His197 were found to participate in protonation/deprotonation of the chromophore accompanying the photoswitching of rsTagRFP fluorescence, whereas residues Met160 and Leu174 were shown to spatially restrict chromophore isomerization, favoring its radiative decay.
Organizational Affiliation:
Synchrotron Radiation Research Section, Macromolecular Crystallography Laboratory, National Cancer Institute, Argonne, IL 60439, USA. pletnevs@mail.nih.gov