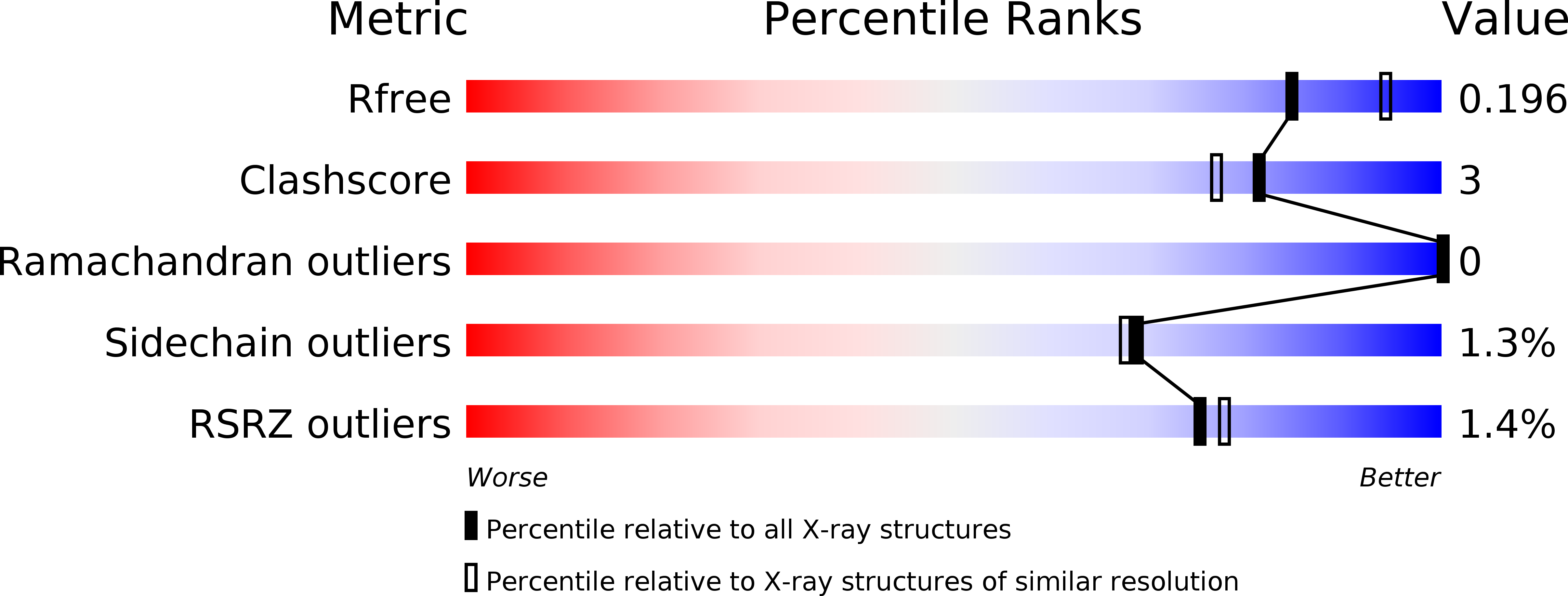
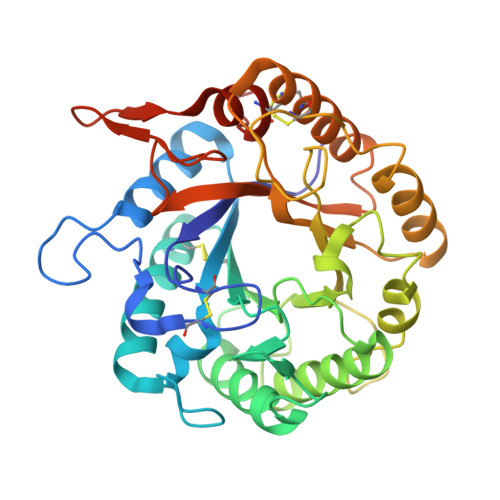
A structural study of Hypocrea jecorina Cel5A.
Lee, T.M., Farrow, M.F., Arnold, F.H., Mayo, S.L.(2011) Protein Sci 20: 1935-1940
- PubMed: 21898652
- DOI: https://doi.org/10.1002/pro.730
- Primary Citation of Related Structures:
3QR3 - PubMed Abstract:
Interest in generating lignocellulosic biofuels through enzymatic hydrolysis continues to rise as nonrenewable fossil fuels are depleted. The high cost of producing cellulases, hydrolytic enzymes that cleave cellulose into fermentable sugars, currently hinders economically viable biofuel production. Here, we report the crystal structure of a prevalent endoglucanase in the biofuels industry, Cel5A from the filamentous fungus Hypocrea jecorina. The structure reveals a general fold resembling that of the closest homolog with a high-resolution structure, Cel5A from Thermoascus aurantiacus. Consistent with previously described endoglucanase structures, the H. jecorina Cel5A active site contains a primarily hydrophobic substrate binding groove and a series of hydrogen bond networks surrounding two catalytic glutamates. The reported structure, however, demonstrates stark differences between side-chain identity, loop regions, and the number of disulfides. Such structural information may aid efforts to improve the stability of this protein for industrial use while maintaining enzymatic activity through revealing nonessential and immutable regions.
Organizational Affiliation:
Biochemistry and Molecular Biophysics Option, California Institute of Technology, Pasadena, California 91125, USA.