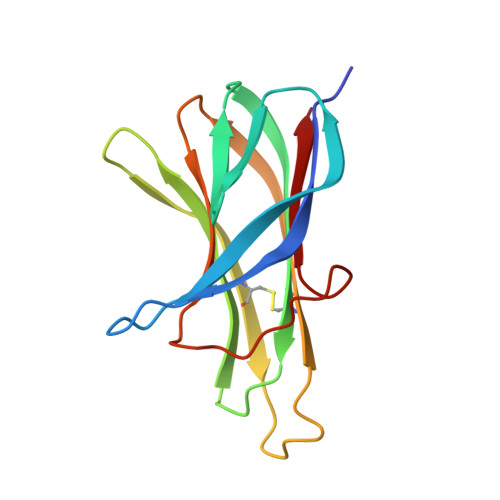
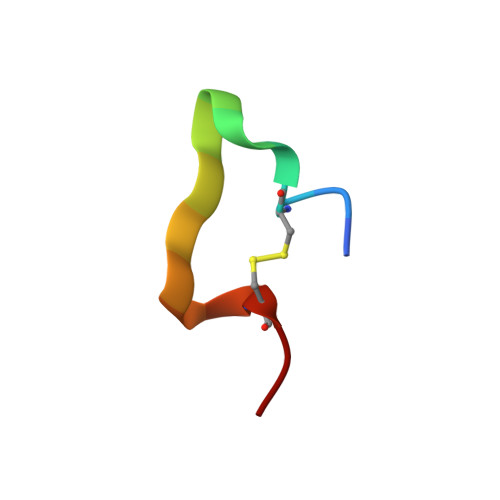
Multiple novel classes of APRIL-specific receptor-blocking peptides isolated by phage display.
Gordon, N.C., Lien, S., Johnson, J., Wallweber, H.J., Tran, T., Currell, B., Mathieu, M., Quan, C., Starovasnik, M.A., Hymowitz, S.G., Kelley, R.F.(2010) J Mol Biol 396: 166-177
- PubMed: 19945466
- DOI: https://doi.org/10.1016/j.jmb.2009.11.041
- Primary Citation of Related Structures:
3K48 - PubMed Abstract:
A proliferation-inducing ligand (APRIL) is a member of the tumor necrosis factor (TNF) ligand superfamily and has a proliferative effect on both normal and tumor cells. The TNF family receptors (B-cell maturation antigen (BCMA), transmembrane activator and CAML-interactor (TACI), and BAFF receptor-3 (BR3)) for APRIL and the closely related ligand, B-cell activating factor of the TNF family (BAFF), bind these ligands through a highly conserved six residue DXL motif ((F/Y/W)-D-X-L-(V/T)-(R/G)). Panning peptide phage display libraries led to the identification of several novel classes of APRIL-binding peptides, which could be grouped by their common sequence motifs. Interestingly, only one of these ten classes consisted of peptides containing the DXL motif. Nevertheless, all classes of peptides prevented APRIL, but not BAFF, from binding BCMA, their shared receptor. Synthetic peptides based on selected sequences inhibited APRIL binding to BCMA with IC(50) values of 0.49-27 microM. An X-ray crystallographic structure of APRIL bound to one of the phage-derived peptides showed that the peptide, lacking the DXL motif, was nevertheless bound in the DXL pocket on APRIL. Our results demonstrate that even though a focused, highly conserved motif is required for APRIL-receptor interaction, remarkably, many novel and distinct classes of peptides are also capable of binding APRIL at the ligand receptor interface.
Organizational Affiliation:
Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080, USA.