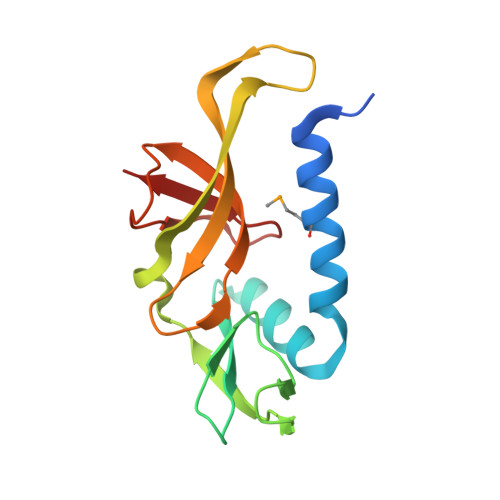
The structure of SSO2064, the first representative of Pfam family PF01796, reveals a novel two-domain zinc-ribbon OB-fold architecture with a potential acyl-CoA-binding role.
Krishna, S.S., Aravind, L., Bakolitsa, C., Caruthers, J., Carlton, D., Miller, M.D., Abdubek, P., Astakhova, T., Axelrod, H.L., Chiu, H.J., Clayton, T., Deller, M.C., Duan, L., Feuerhelm, J., Grant, J.C., Han, G.W., Jaroszewski, L., Jin, K.K., Klock, H.E., Knuth, M.W., Kumar, A., Marciano, D., McMullan, D., Morse, A.T., Nigoghossian, E., Okach, L., Reyes, R., Rife, C.L., van den Bedem, H., Weekes, D., Xu, Q., Hodgson, K.O., Wooley, J., Elsliger, M.A., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1160-1166
- PubMed: 20944206
- DOI: https://doi.org/10.1107/S1744309110002514
- Primary Citation of Related Structures:
3IRB - PubMed Abstract:
SSO2064 is the first structural representative of PF01796 (DUF35), a large prokaryotic family with a wide phylogenetic distribution. The structure reveals a novel two-domain architecture comprising an N-terminal, rubredoxin-like, zinc ribbon and a C-terminal, oligonucleotide/oligosaccharide-binding (OB) fold domain. Additional N-terminal helical segments may be involved in protein-protein interactions. Domain architectures, genomic context analysis and functional evidence from certain bacterial representatives of this family suggest that these proteins form a novel fatty-acid-binding component that is involved in the biosynthesis of lipids and polyketide antibiotics and that they possibly function as acyl-CoA-binding proteins. This structure has led to a re-evaluation of the DUF35 family, which has now been split into two entries in the latest Pfam release (v.24.0).
Organizational Affiliation:
Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, Menlo Park,California, USA.