Myosin VI undergoes cargo-mediated dimerization
Yu, C., Feng, W., Wei, Z., Miyanoiri, Y., Wen, W., Zhao, Y., Zhang, M.(2009) Cell 138: 537-548
- PubMed: 19665975
- DOI: https://doi.org/10.1016/j.cell.2009.05.030
- Primary Citation of Related Structures:
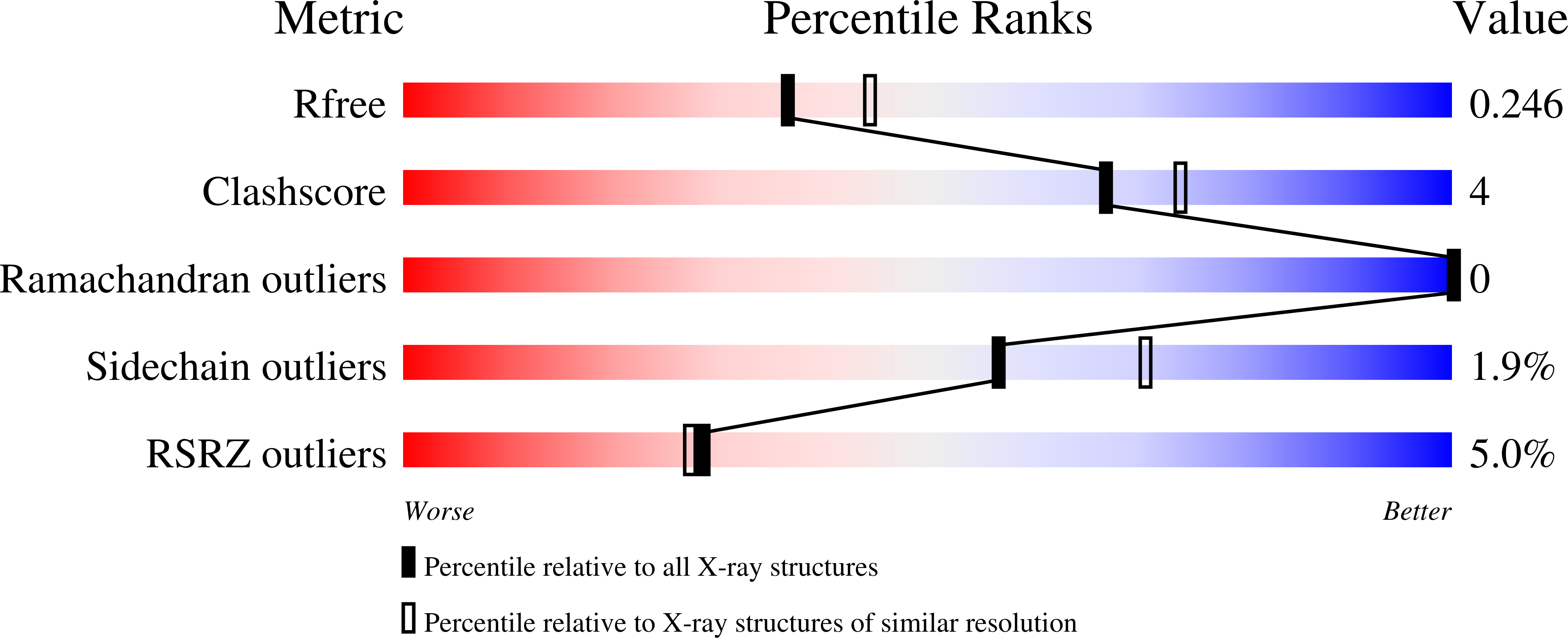
2KIA, 3H8D - PubMed Abstract:
Myosin VI is the only known molecular motor that moves toward the minus ends of actin filaments; thus, it plays unique roles in diverse cellular processes. The processive walking of myosin VI on actin filaments requires dimerization of the motor, but the protein can also function as a nonprocessive monomer. The molecular mechanism governing the monomer-dimer conversion is not clear. We report the high-resolution NMR structure of the cargo-free myosin VI cargo-binding domain (CBD) and show that it is a stable monomer in solution. The myosin VI CBD binds to a fragment of the clathrin-coated vesicle adaptor Dab2 with a high affinity, and the X-ray structure of the myosin VI CBD in complex with Dab2 reveals that the motor undergoes a cargo-binding-mediated dimerization. The cargo-binding-induced dimerization may represent a general paradigm for the regulation of processivity for myosin VI as well as other myosins, including myosin VII and myosin X.
Organizational Affiliation:
Department of Biochemistry, Molecular Neuroscience Center, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong.