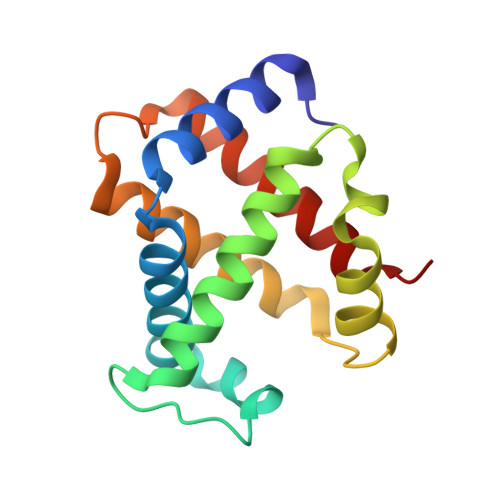
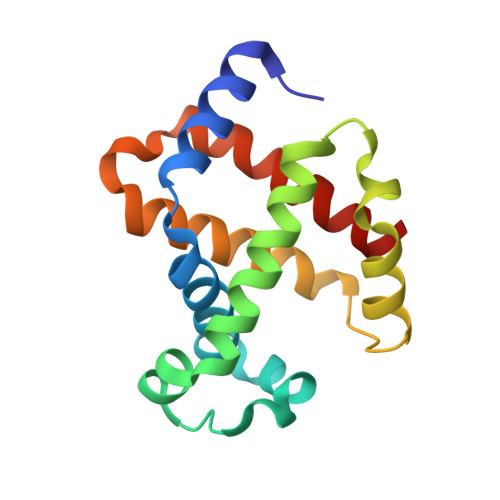
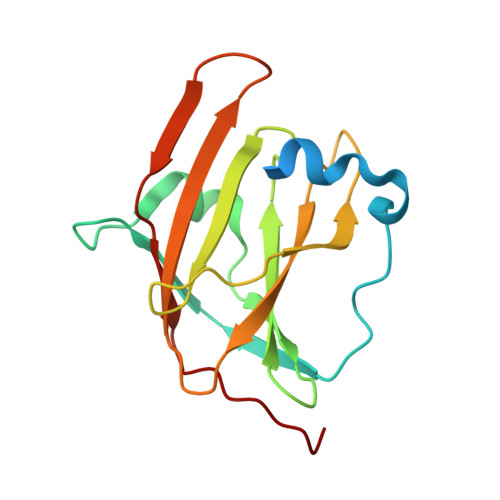
Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein, IsdH
Krishna Kumar, K., Jacques, D.A., Pishchany, G., Caradoc-Davies, T., Spirig, T., Malmirchegini, G.R., Langley, D.B., Dickson, C.F., Mackay, J.P., Clubb, R.T., Skaar, E.P., Guss, J.M., Gell, D.A.(2011) J Biol Chem 286: 38439-38447
- PubMed: 21917915
- DOI: https://doi.org/10.1074/jbc.M111.287300
- Primary Citation of Related Structures:
3SZK - PubMed Abstract:
Pathogens must steal iron from their hosts to establish infection. In mammals, hemoglobin (Hb) represents the largest reservoir of iron, and pathogens express Hb-binding proteins to access this source. Here, we show how one of the commonest and most significant human pathogens, Staphylococcus aureus, captures Hb as the first step of an iron-scavenging pathway. The x-ray crystal structure of Hb bound to a domain from the Isd (iron-regulated surface determinant) protein, IsdH, is the first structure of a Hb capture complex to be determined. Surface mutations in Hb that reduce binding to the Hb-receptor limit the capacity of S. aureus to utilize Hb as an iron source, suggesting that Hb sequence is a factor in host susceptibility to infection. The demonstration that pathogens make highly specific recognition complexes with Hb raises the possibility of developing inhibitors of Hb binding as antibacterial agents.
Organizational Affiliation:
School of Molecular Bioscience, University of Sydney, New South Wales 2006, Australia.