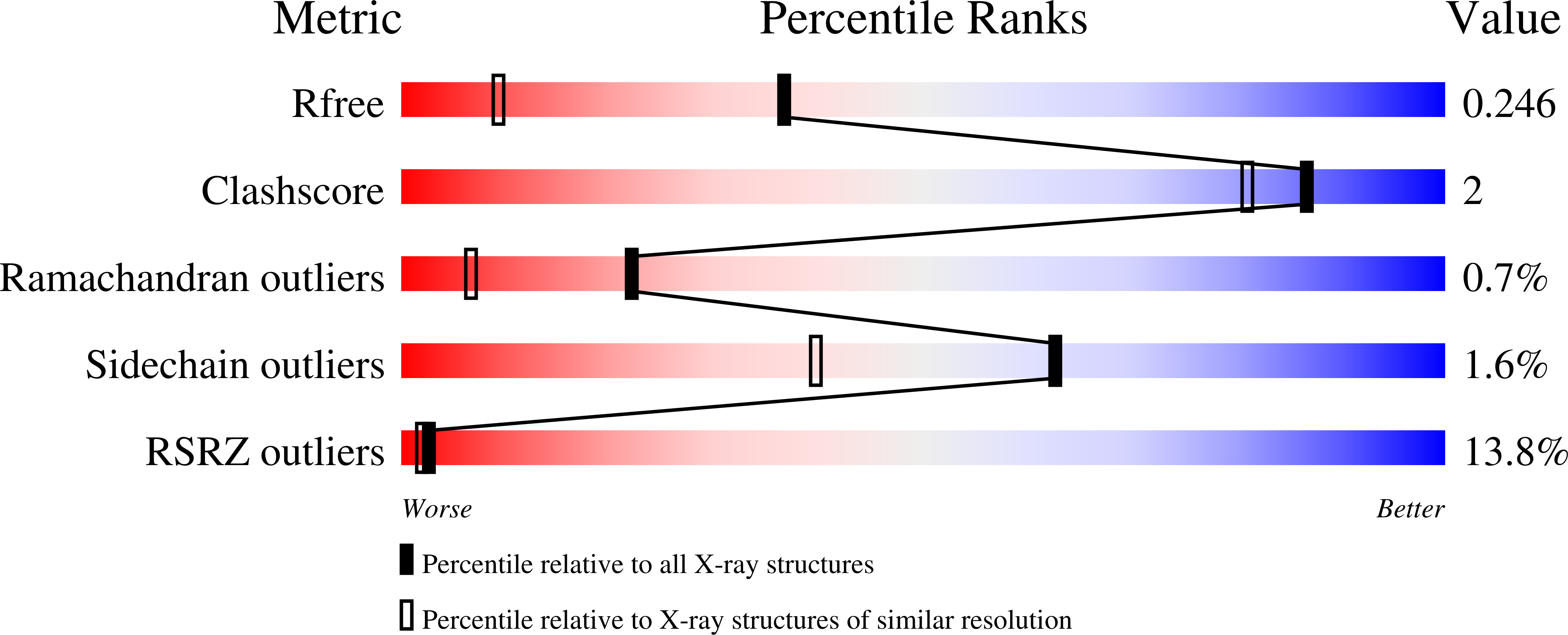
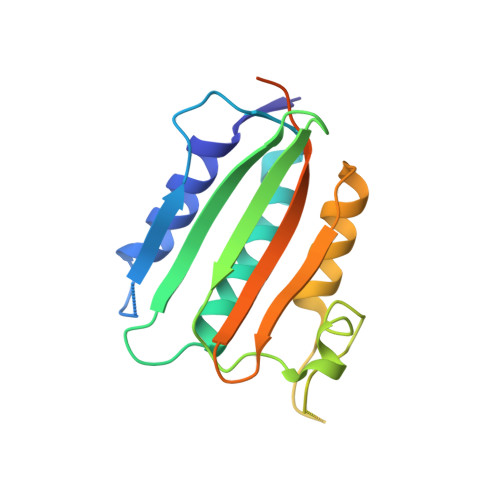
ATP forms a stable complex with the essential histidine kinase WalK (YycG) domain.
Celikel, R., Veldore, V.H., Mathews, I., Devine, K.M., Varughese, K.I.(2012) Acta Crystallogr D Biol Crystallogr 68: 839-845
- PubMed: 22751669
- DOI: https://doi.org/10.1107/S090744491201373X
- Primary Citation of Related Structures:
3SL2 - PubMed Abstract:
In Bacillus subtilis, the WalRK (YycFG) two-component system coordinates murein synthesis with cell division. It regulates the expression of autolysins that function in cell-wall remodeling and of proteins that modulate autolysin activity. The transcription factor WalR is activated upon phosphorylation by the histidine kinase WalK, a multi-domain homodimer. It autophosphorylates one of its histidine residues by transferring the γ-phosphate from ATP bound to its ATP-binding domain. Here, the high-resolution crystal structure of the ATP-binding domain of WalK in complex with ATP is presented at 1.61 Å resolution. The bound ATP remains intact in the crystal lattice. It appears that the strong binding interactions and the nature of the binding pocket contribute to its stability. The triphosphate moiety of ATP wraps around an Mg(2+) ion, providing three O atoms for coordination in a near-ideal octahedral geometry. The ATP molecule also makes strong interactions with the protein. In addition, there is a short contact between the exocyclic O3' of the sugar ring and O2B of the β-phosphate, implying an internal hydrogen bond. The stability of the WalK-ATP complex in the crystal lattice suggests that such a complex may exist in vivo poised for initiation of signal transmission. This feature may therefore be part of the sensing mechanism by which the WalRK two-component system is so rapidly activated when cells encounter conditions conducive for growth.
Organizational Affiliation:
Department of Physiology and Biophysics, University of Arkansas for Medical Sciences, 4301 West Markham Street, Little Rock, AR 72205, USA.