Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency.
Nishio, M., Kamiya, Y., Mizushima, T., Wakatsuki, S., Sasakawa, H., Yamamoto, K., Uchiyama, S., Noda, M., McKay, A.R., Fukui, K., Hauri, H.P., Kato, K.(2010) Proc Natl Acad Sci U S A 107: 4034-4039
- PubMed: 20142513
- DOI: https://doi.org/10.1073/pnas.0908526107
- Primary Citation of Related Structures:
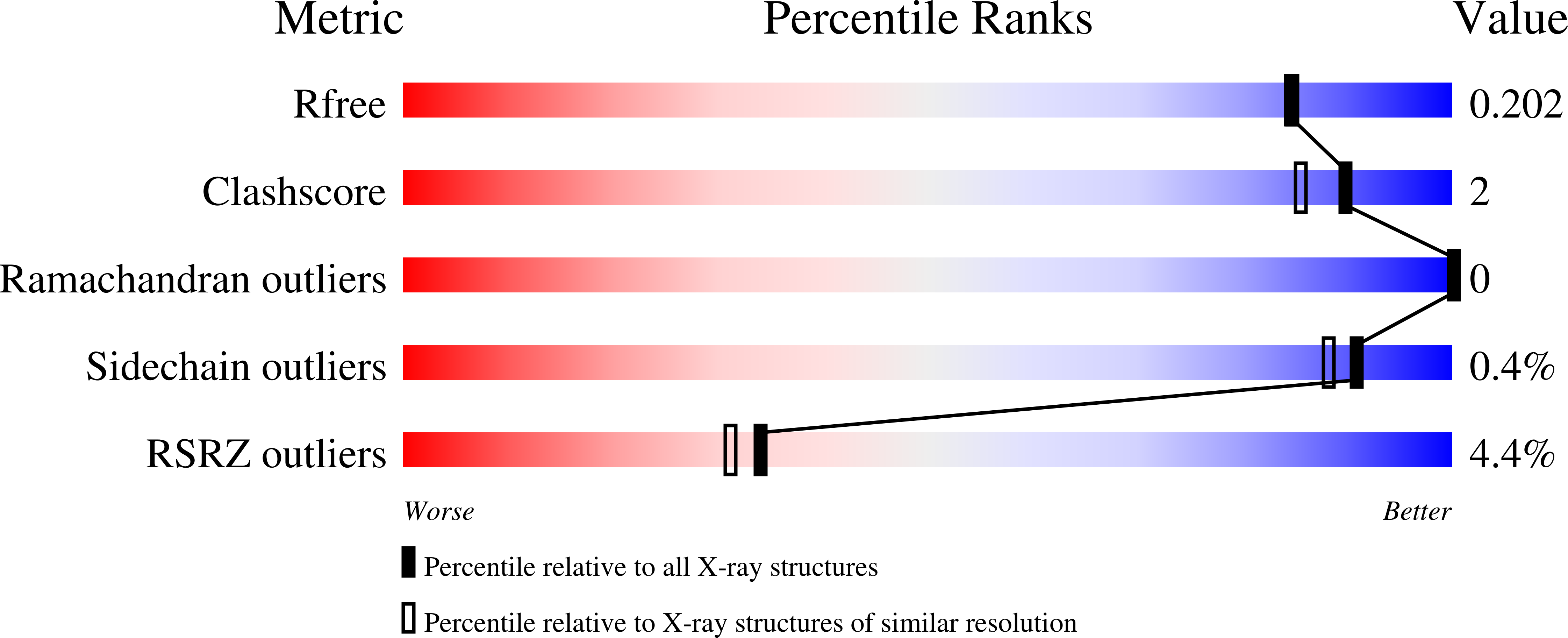
3A4U - PubMed Abstract:
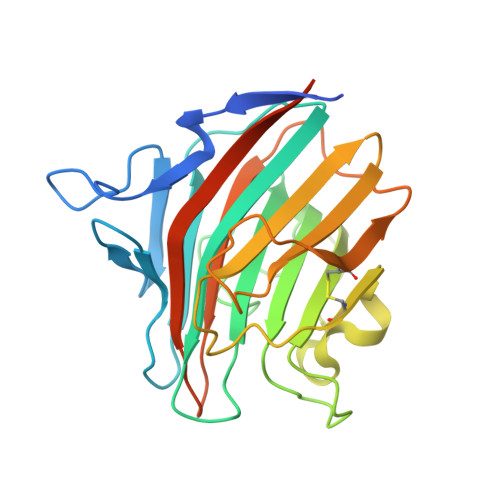
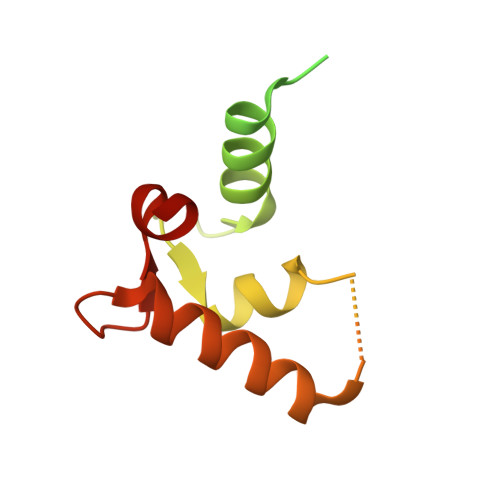
Combined deficiency of coagulation factors V and VIII (F5F8D), an autosomal recessive disorder characterized by coordinate reduction in the plasma levels of factor V (FV) and factor VIII (FVIII), is genetically linked to mutations in the transmembrane lectin ERGIC-53 and the soluble calcium-binding protein MCFD2. Growing evidence indicates that these two proteins form a complex recycling between the endoplasmic reticulum (ER) and the ER-Golgi intermediate compartment and thereby function as a cargo receptor in the early secretory pathway of FV and FVIII. For better understanding of the mechanisms underlying the functional coordination of ERGIC-53 and MCFD2, we herein characterize their interaction by x-ray crystallographic analysis in conjunction with NMR and ultracentrifugation analyses. Inspection of the combined data reveals that ERGIC-53-CRD binds MCFD2 through its molecular surface remote from the sugar-binding site, giving rise to a 11 complex in solution. The interaction is independent of sugar-binding of ERGIC-53 and involves most of the missense mutation sites of MCFD2 so far reported in F5F8D. Comparison with the previously reported uncomplexed structure of each protein indicates that MCFD2 but not ERGIC-53-CRD undergoes significant conformational alterations upon complex formation. Our findings provide a structural basis for the cooperative interplay between ERGIC-53 and MCFD2 in capturing FV and FVIII.
Organizational Affiliation:
Department of Structural Biology and Biomolecular Engineering, Graduate School of Pharmaceutical Sciences, Nagoya City University, 3-1 Tanabe-dori, Mizuho-ku, Nagoya 467-8603, Japan.