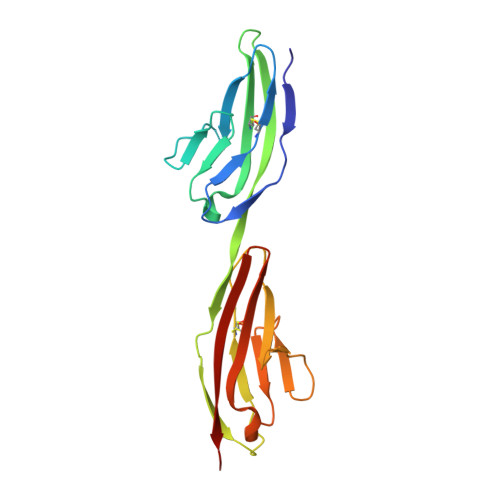
Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Kulahin, N., Kristensen, O., Rasmussen, K.K., Olsen, L., Rydberg, P., Vestergaard, B., Kastrup, J.S., Berezin, V., Bock, E., Walmod, P.S., Gajhede, M.(2011) Structure 19: 203
- PubMed: 21300289
- DOI: https://doi.org/10.1016/j.str.2010.12.014
- Primary Citation of Related Structures:
2JLL, 2V5T, 2WIM, 2XY1, 2XY2, 2XYC - PubMed Abstract:
The ectodomain of olfactory cell adhesion molecule (OCAM/NCAM2/RNCAM) consists of five immunoglobulin (Ig) domains (IgI-V), followed by two fibronectin-type 3 (Fn3) domains (Fn3I-II). A complete structural model of the entire ectodomain of human OCAM has been assembled from crystal structures of six recombinant proteins corresponding to different regions of the ectodomain. The model is the longest experimentally based composite structural model of an entire IgCAM ectodomain. It displays an essentially linear arrangement of IgI-V, followed by bends between IgV and Fn3I and between Fn3I and Fn3II. Proteins containing IgI-IgII domains formed stable homodimers in solution and in crystals. Dimerization could be disrupted in vitro by mutations in the dimer interface region. In conjunction with the bent ectodomain conformation, which can position IgI-V parallel with the cell surface, the IgI-IgII dimerization enables OCAM-mediated trans-interactions with an intercellular distance of about 20 nm, which is consistent with that observed in synapses.
Organizational Affiliation:
Protein Laboratory, Department of Neuroscience and Pharmacology, Faculty of Health Sciences, University of Copenhagen, DK-2200 Copenhagen N, Denmark.