Solution Structure of Polytheonamide B, a Highly Cytotoxic Nonribosomal Polypeptide from Marine Sponge
Hamada, T., Matsunaga, S., Fujiwara, M., Fujita, K., Hirota, H., Schmucki, R., Guntert, P., Fusetani, N.(2010) J Am Chem Soc
- PubMed: 20795624
- DOI: https://doi.org/10.1021/ja104616z
- Primary Citation of Related Structures:
2RQO - PubMed Abstract:
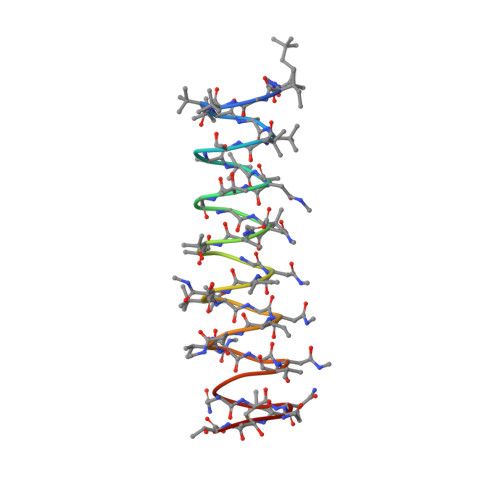
Polytheonamide B (pTB), a highly cytotoxic polypeptide, is one of the most unusual nonribosomal peptides of sponge origin. pTB is a linear 48-residue peptide with alternating D- and L-amino acids and contains a total of eight types of nonproteinogenic amino acids. To investigate the mechanisms underlying its cytotoxic activity, we determined the three-dimensional structure of pTB by NMR spectroscopy, structure calculation, and energy minimization. pTB adopts a single right-handed β(6.3)-helical structure in a 1:1 mixture of methanol/chloroform with a length of approximately 45 A and a hydrophilic pore of ca. 4 A inner diameter. These features indicate that pTB molecules form transmembrane channels that permeate monovalent cations as gramicidin A channels do. The strong cytotoxicity of pTB can be ascribed to its ability to form single molecule channels through biological membranes.
Organizational Affiliation:
Laboratory of Aquatic Natural Products Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo 113-8657, Japan. thamada@sci.kagoshima-u.ac.jp