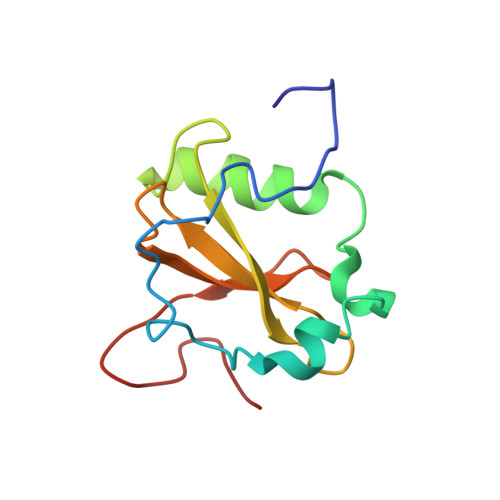
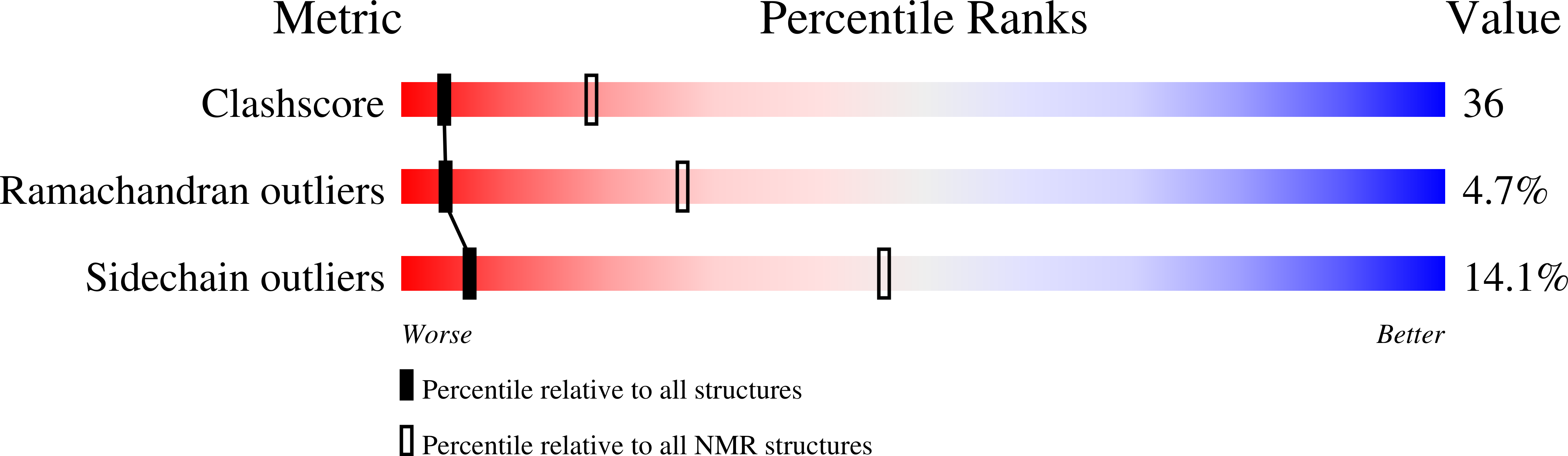Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Manuse, S., Jean, N.L., Guinot, M., Lavergne, J.P., Laguri, C., Bougault, C.M., VanNieuwenhze, M.S., Grangeasse, C., Simorre, J.P.(2016) Nat Commun 7: 12071-12071
- PubMed: 27346279
- DOI: https://doi.org/10.1038/ncomms12071
- Primary Citation of Related Structures:
2ND9, 2NDA - PubMed Abstract:
Accurate placement of the bacterial division site is a prerequisite for the generation of two viable and identical daughter cells. In Streptococcus pneumoniae, the positive regulatory mechanism involving the membrane protein MapZ positions precisely the conserved cell division protein FtsZ at the cell centre. Here we characterize the structure of the extracellular domain of MapZ and show that it displays a bi-modular structure composed of two subdomains separated by a flexible serine-rich linker. We further demonstrate in vivo that the N-terminal subdomain serves as a pedestal for the C-terminal subdomain, which determines the ability of MapZ to mark the division site. The C-terminal subdomain displays a patch of conserved amino acids and we show that this patch defines a structural motif crucial for MapZ function. Altogether, this structure-function analysis of MapZ provides the first molecular characterization of a positive regulatory process of bacterial cell division.
Organizational Affiliation:
CNRS, Molecular Microbiology and Structural Biochemistry, UMR 5086, 7 passage du Vercors, Lyon 69367, France.