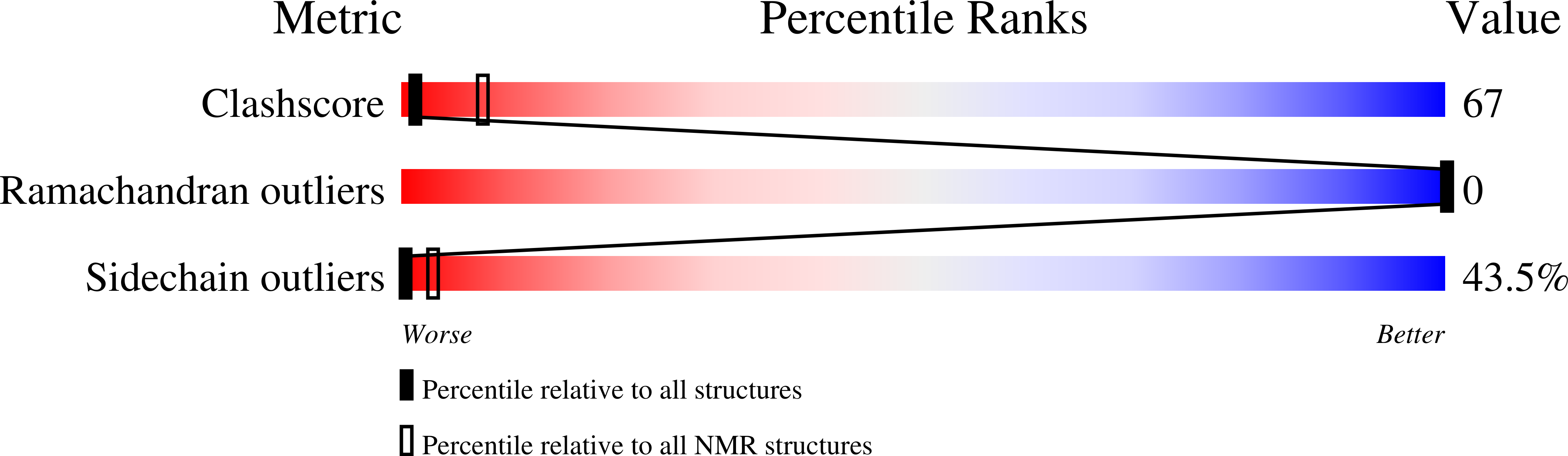Oligomerization of the antimicrobial peptide Protegrin-5 in a membrane-mimicking environment. Structural studies by high-resolution NMR spectroscopy.
Usachev, K.S., Kolosova, O.A., Klochkova, E.A., Yulmetov, A.R., Aganov, A.V., Klochkov, V.V.(2017) Eur Biophys J 46: 293-300
- PubMed: 27589857
- DOI: https://doi.org/10.1007/s00249-016-1167-5
- Primary Citation of Related Structures:
2NC7 - PubMed Abstract:
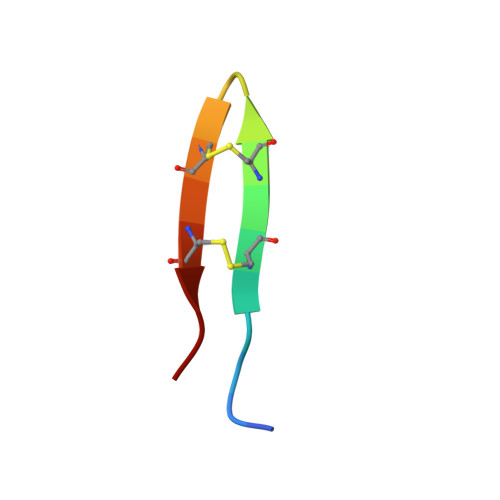
Protegrin pore formation is believed to occur in a stepwise fashion that begins with a nonspecific peptide interaction with the negatively charged bacterial cell walls via hydrophobic and positively charged amphipathic surfaces. There are five known nature protegrins (PG1-PG5), and early studies of PG-1 (PDB ID:1PG1) shown that it could form antiparallel dimer in membrane mimicking environment which could be a first step for further oligomeric membrane pore formation. Later, we solved PG-2 (PDB ID:2MUH) and PG-3 (PDB ID:2MZ6) structures in the same environment and for PG-3 observed a strong d αα NOE effects between residues R18 and F12, V14, and V16. These "inconsistent" with monomer structure NOEs appears due to formation of an additional antiparallel β-sheet between two monomers. It was also suggested that there is a possible association of protegrins dimers to form octameric or decameric β-barrels in an oligomer state. In order to investigate a more detailed oligomerization process of protegrins, in the present article we report the monomer (PDB ID: 2NC7) and octamer pore structures of the protegrin-5 (PG-5) in the presence of DPC micelles studied by solution NMR spectroscopy. In contrast to PG-1, PG-2, and PG-3 studies, for PG-5 we observed not only dimer NOEs but also several additional NOEs between side chains, which allows us to calculate an octamer pore structure of PG-5 that was in good agreement with previous AFM and PMF data.
Organizational Affiliation:
Kazan Federal University, 18 Kremlevskaya St., 420008, Kazan, Russian Federation.