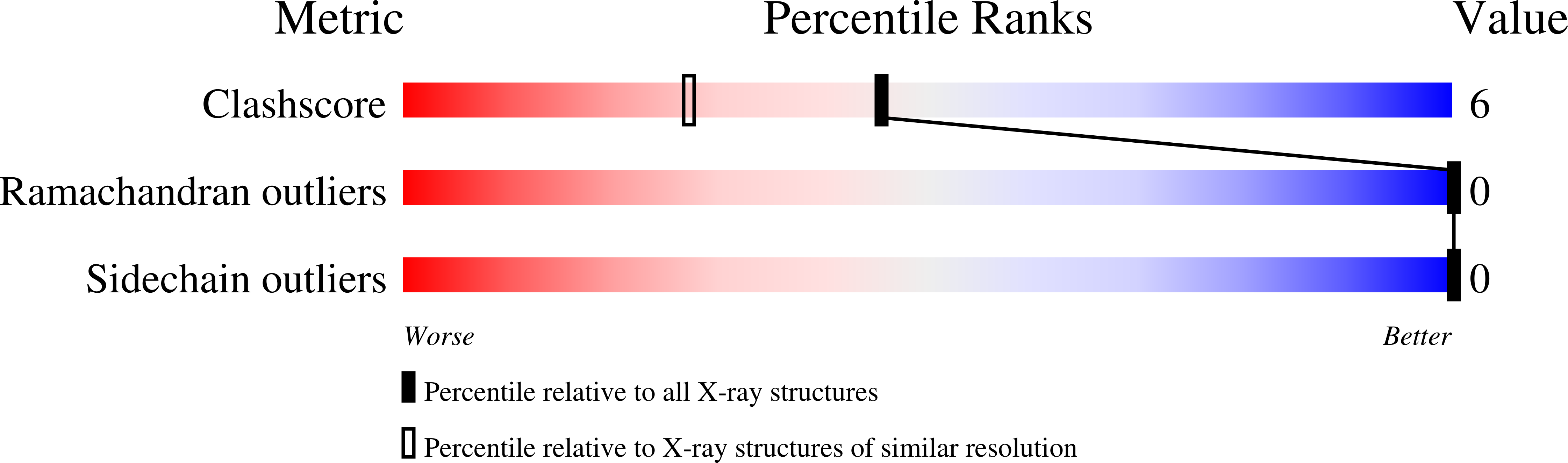High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)(4)-Xaa-Yaa-Gly-(Pro-Pro-Gly)(4)]: Implications for triple-helix hydration and Hyp(X) puckering.
Okuyama, K., Hongo, C., Wu, G., Mizuno, K., Noguchi, K., Ebisuzaki, S., Tanaka, Y., Nishino, N., Bachinger, H.P.(2009) Biopolymers 91: 361-372
- PubMed: 19137577
- DOI: https://doi.org/10.1002/bip.21138
- Primary Citation of Related Structures:
2D3F, 2D3H - PubMed Abstract:
Structures of (Pro-Pro-Gly)4-Xaa-Yaa-Gly-(Pro-Pro-Gly)4 (ppg9-XYG) where (Xaa, Yaa)=(Pro, Hyp), (Hyp, Pro) or (Hyp, Hyp) were analyzed at high resolution using synchrotron radiation. Molecular and crystal structures of these peptides are very similar to those of the (Pro-Pro-Gly)9 peptide. The results obtained in this study, together with those obtained from related compounds, indicated the puckering propensity of the Hyp in the X position: (1) Hyp(X) residues involved in the Hyp(X):Pro(Y) stacking pairs prefer the down-puckering conformation, as in ppg9-OPG, and ppg9-OOG; (2) Hyp(X) residues involved in the Hyp(X):Hyp(Y) stacking pairs prefer the up-puckering conformation if there is no specific reason to adopt the down-puckering conformation. Water molecules in these peptide crystals are classified into two groups, the 1st and 2nd hydration waters. Water molecules in the 1st hydration group have direct hydrogen bonds with peptide oxygen atoms, whereas those in the 2nd hydration group do not. Compared with globular proteins, the number of water molecules in the 2nd hydration shell of the ppg9-XYG peptides is very large, likely due to the unique rod-like molecular structure of collagen model peptides. In the collagen helix, the amino acid residues in the X and Y positions must protrude outside of the triple helix, which forces even the hydrophobic side chains, such as Pro, to be exposed to the surrounding water molecules. Therefore, most of the waters in the 2nd hydration shell are covering hydrophobic Pro side chains by forming clathrate structures.
Organizational Affiliation:
Department of Macromolecular Science, Graduate School of Science, Osaka University, Toyonaka, Osaka 560-0043, Japan. okuyamak@chem.sci.osaka-u.ac.jp