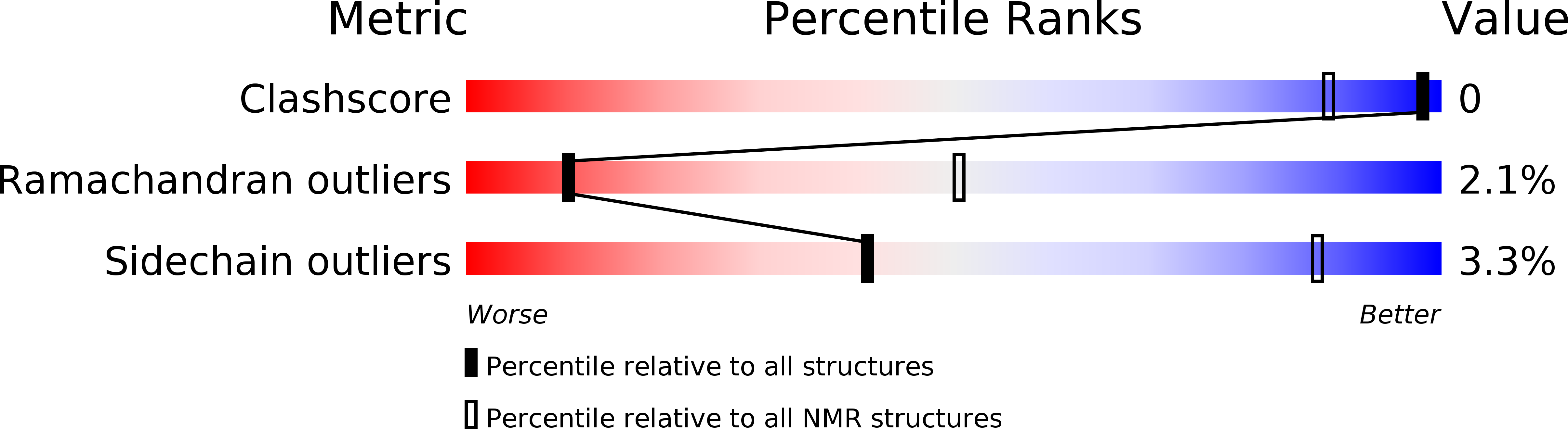Path to Collagenolysis: COLLAGEN V TRIPLE-HELIX MODEL BOUND PRODUCTIVELY AND IN ENCOUNTERS BY MATRIX METALLOPROTEINASE-12.
Prior, S.H., Byrne, T.S., Tokmina-Roszyk, D., Fields, G.B., Van Doren, S.R.(2016) J Biol Chem 291: 7888-7901
- PubMed: 26887942
- DOI: https://doi.org/10.1074/jbc.M115.703124
- Primary Citation of Related Structures:
2N8R - PubMed Abstract:
Collagenolysis is essential in extracellular matrix homeostasis, but its structural basis has long been shrouded in mystery. We have developed a novel docking strategy guided by paramagnetic NMR that positions a triple-helical collagen V mimic (synthesized with nitroxide spin labels) in the active site of the catalytic domain of matrix metalloproteinase-12 (MMP-12 or macrophage metalloelastase) primed for catalysis. The collagenolytically productive complex forms by utilizing seven distinct subsites that traverse the entire length of the active site. These subsites bury ∼1,080 Å(2)of surface area, over half of which is contributed by the trailing strand of the synthetic collagen V mimic, which also appears to ligate the catalytic zinc through the glycine carbonyl oxygen of its scissile G∼VV triplet. Notably, the middle strand also occupies the full length of the active site where it contributes extensive interfacial contacts with five subsites. This work identifies, for the first time, the productive and specific interactions of a collagen triple helix with an MMP catalytic site. The results uniquely demonstrate that the active site of the MMPs is wide enough to accommodate two strands from collagen triple helices. Paramagnetic relaxation enhancements also reveal an extensive array of encounter complexes that form over a large part of the catalytic domain. These transient complexes could possibly facilitate the formation of collagenolytically active complexes via directional Brownian tumbling.
Organizational Affiliation:
From the Department of Biochemistry, University of Missouri, Columbia, Missouri 65211.