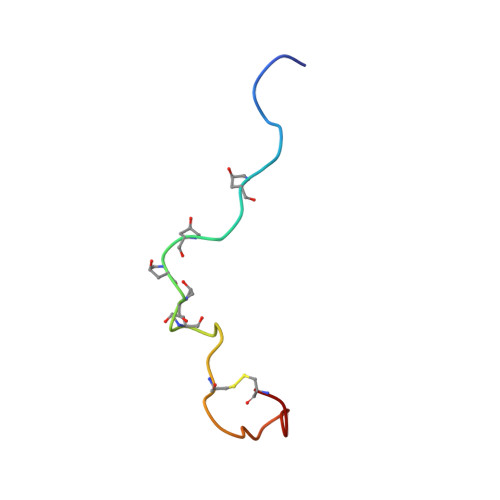
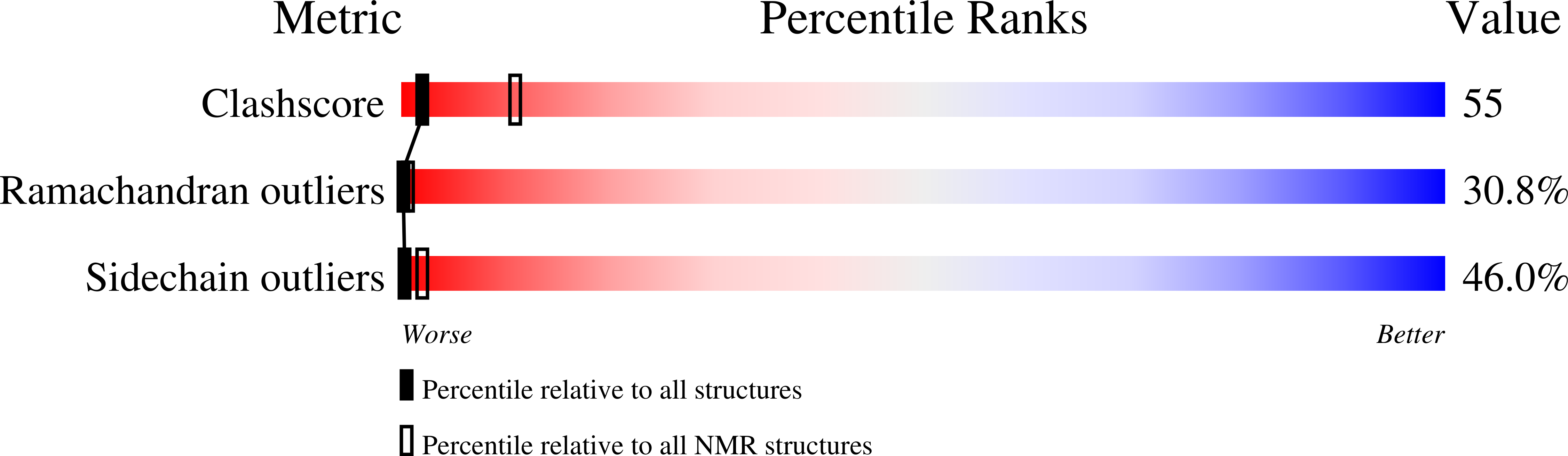NMR studies of the solution conformation of the sex peptide from Drosophila melanogaster.
Moehle, K., Freund, A., Kubli, E., Robinson, J.A.(2011) FEBS Lett 585: 1197-1202
- PubMed: 21439282
- DOI: https://doi.org/10.1016/j.febslet.2011.03.040
- Primary Citation of Related Structures:
2LAQ - PubMed Abstract:
The insect sex peptide (SP) elicits a variety of biological responses upon transfer to the mated female. SP contains 36 amino acids, including a tryptophan-rich N-terminal region, a central region containing five hydroxyproline (Hyp) residues, and a C-terminal region enclosed by a disulfide bridge. The solution structure of SP, studied here using NMR spectroscopy, includes a motif WPWN that adopts a type I β-turn in the N-terminal Trp-rich region. This turn region is connected to the central Hyp-rich region, which adopts extended and/or PPII-like conformations. The C-terminal disulfide-bonded loop populates helical turns or nascent helical structure. Overall, the results reveal a rather flexible peptide that lacks a compact folded structure in solution.
Organizational Affiliation:
Chemistry Department, University of Zurich, Winterthurerstrasse 190, 8057 Zurich, Switzerland.