Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2
Li, H., Fung, K.L., Jin, D.Y., Chung, S.S., Ching, Y.P., Ng, I.O., Sze, K.H., Ko, B.C., Sun, H.(2007) Proteins 67: 1154-1166
- PubMed: 17380510
- DOI: https://doi.org/10.1002/prot.21361
- Primary Citation of Related Structures:
2H80 - PubMed Abstract:
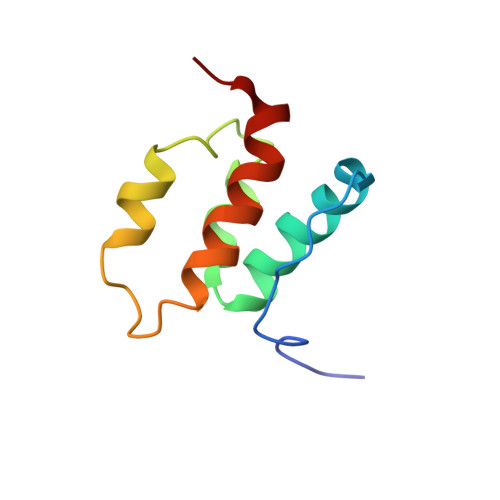
The deleted in liver cancer 2 (DLC2) is a tumor suppressor gene, frequently found to be underexpressed in hepatocellular carcinoma. DLC2 is a multidomain protein containing a sterile alpha-motif (SAM) domain, a GTPase-activating protein (GAP) domain, and a lipid-binding StAR-related lipid-transfer (START) domain. The SAM domain of DLC2, DLC2-SAM, exhibits a low level of sequence homology (15-30%) with other SAM domains, and appears to be the prototype of a new subfamily of SAM domains found in DLC2-related proteins. In the present study, we have determined the three-dimensional solution structure of DLC2-SAM using NMR methods together with molecular dynamics simulated annealing. In addition, we performed a backbone dynamics study. The DLC2-SAM packed as a unique four alpha-helical bundle stabilized by interhelix hydrophobic interactions. The arrangement of the four helices is distinct from all other known SAM domains. In contrast to some members of the SAM domain family which form either dimers or oligomers, both biochemical analyses and rotational correlation time (tau(c)) measured by backbone 15N relaxation experiments indicated that DLC2-SAM exists as a monomer in solution. The interaction of DLC2-SAM domain with sodium dodecyl sulfate (SDS) micelles and 1,2-dimyristoyl-sn-glycerol-3-phosphatidylglycerol (DMPG) phospholipids was examined by CD and NMR spectroscopic techniques. The DLC2-SAM exhibits membrane binding properties accompanied by minor loss of the secondary structure of the protein. Deletion studies showed that the self-association of DLC2 in vivo does not require SAM domain, instead, a protein domain consisting of residues 120-672 mediates the self-association of DLC2.
Organizational Affiliation:
Department of Chemistry and Open Laboratory of Chemical Biology, The University of Hong Kong, Pokfulam, Hong Kong, People's Republic of China.