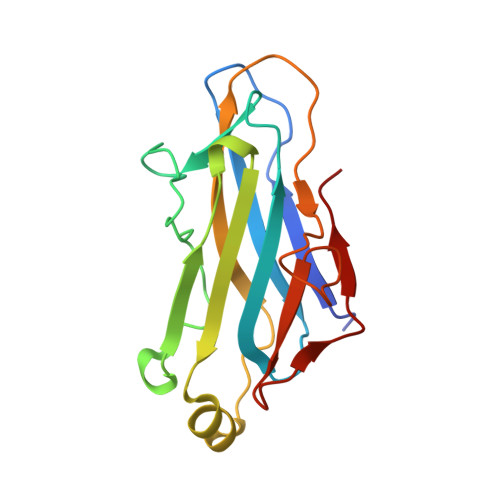
Insights Into the Structural Determinants of Cohesin-Dockerin Specificity Revealed by the Crystal Structure of the Type II Cohesin from Clostridium Thermocellum Sdba.
Carvalho, A.L., Pires, V.M.R., Gloster, T.M., Turkenburg, J.P., Prates, J.A.M., Ferreira, L.M.A., Romao, M.J., Davies, G.J., Fontes, C.M.G.A., Gilbert, H.J.(2005) J Mol Biol 349: 909
- PubMed: 15913653
- DOI: https://doi.org/10.1016/j.jmb.2005.04.037
- Primary Citation of Related Structures:
2BM3 - PubMed Abstract:
The plant cell wall degrading enzymes expressed by anaerobic microorganisms form large multienzyme complexes (cellulosomes). Cellulosomes assemble by the Type I dockerins on the catalytic subunits binding to the reiterated Type I cohesins in the molecular scaffold, while Type II dockerin-cohesin interactions anchor the complex onto the bacterial cell surface. Type I and Type II cohesin, dockerin pairs show no cross-specificity. Here we report the crystal structure of the Type II cohesin (CohII) from the Clostridium thermocellum cell surface anchoring protein SdbA. The protein domain contains nine beta-strands and a small alpha-helix. The beta-strands assemble into two elongated beta-sheets that display a typical jelly roll fold. The structure of CohII is very similar to Type I cohesins, and the dockerin binding site, which is centred at beta-strands 3, 5 and 6, is likely to be conserved in the two proteins. Subtle differences in the topology of the binding sites and a lack of sequence identity in the beta-strands that comprise the core of the dockerin binding site explain why Type I and Type II cohesins display such distinct specificities for their target dockerins.
Organizational Affiliation:
REQUIMTE/CQFB, Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, 2829-516 Caparica, Portugal.