Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
Tremblay, L.W., Zhang, G., Dai, J., Dunaway-Mariano, D., Allen, K.N.(2005) J Am Chem Soc 127: 5298-5299
- PubMed: 15826149
- DOI: https://doi.org/10.1021/ja0509073
- Primary Citation of Related Structures:
1Z4N, 1Z4O - PubMed Abstract:
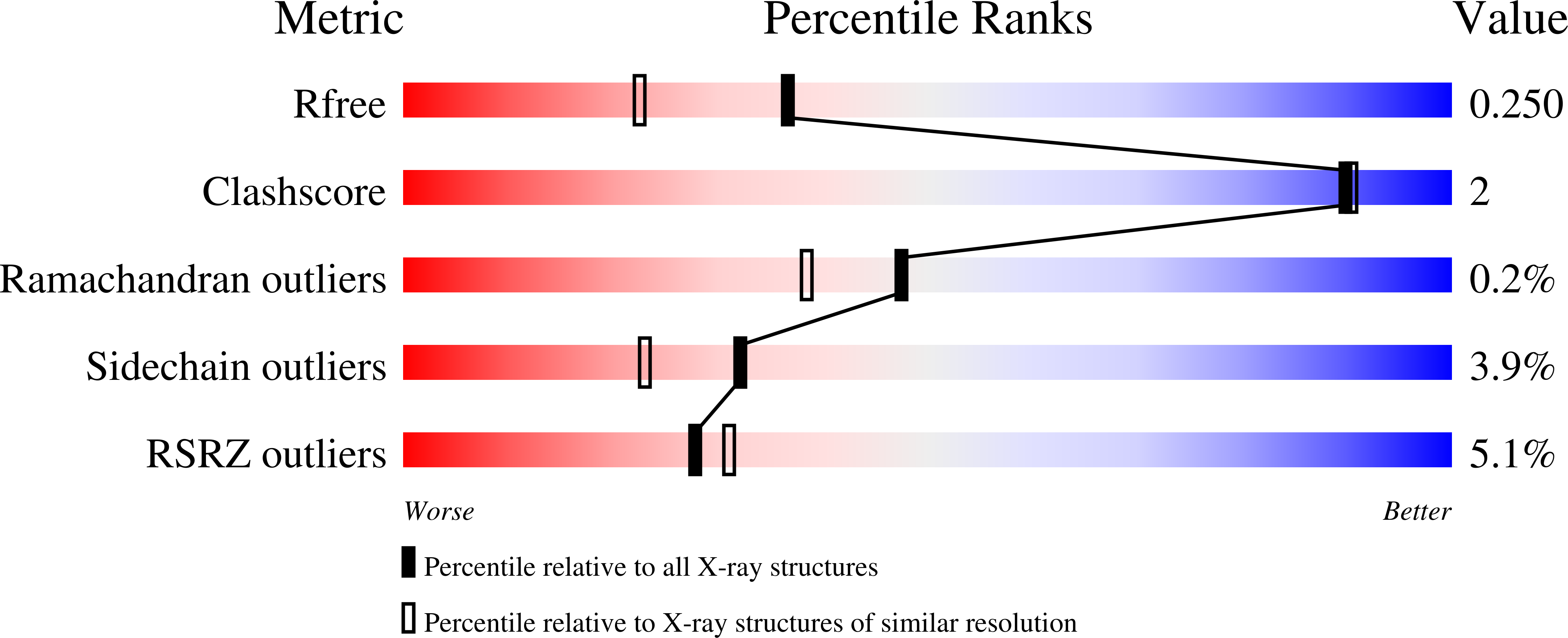
This communication reports the X-ray crystal structure of the alpha-d-galactose-1-phosphate complex with that of Lactococcus lactis beta-phosphoglucomutase (beta-PGM) crystallized in the presence of Mg2+ cofactor and the enzyme-to-phosphorus ratio determined by protein and phosphate analyses of the crystalline complex. The 1:1 ratio determined for this complex was compared to the 1:2 ratio determined for the crystals of beta-PGM grown in the presence of substrate and Mg2+ cofactor. This result verifies the published structure assignment of this latter complex as the phosphorane adduct formed by covalent bonding between the active site Asp8 carboxylate to the C(1)phosphorus of the beta-glucose 1,6-bisphosphate ligand and rules out the proposal of a beta-PGM-glucose-6-phosphate-1-MgF3- complex.
Organizational Affiliation:
Department of Physiology and Biophysics, Boston University School of Medicine, Boston, Massachusetts 02118-2394, USA.