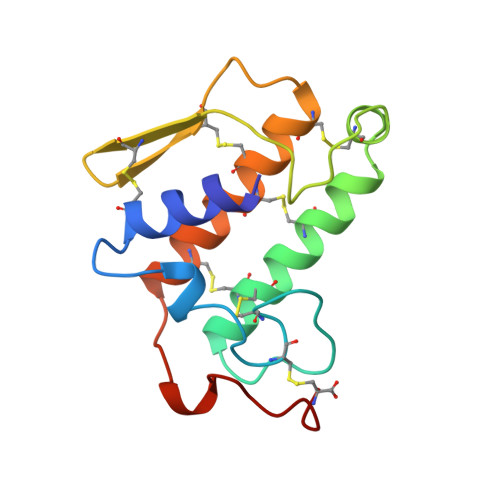
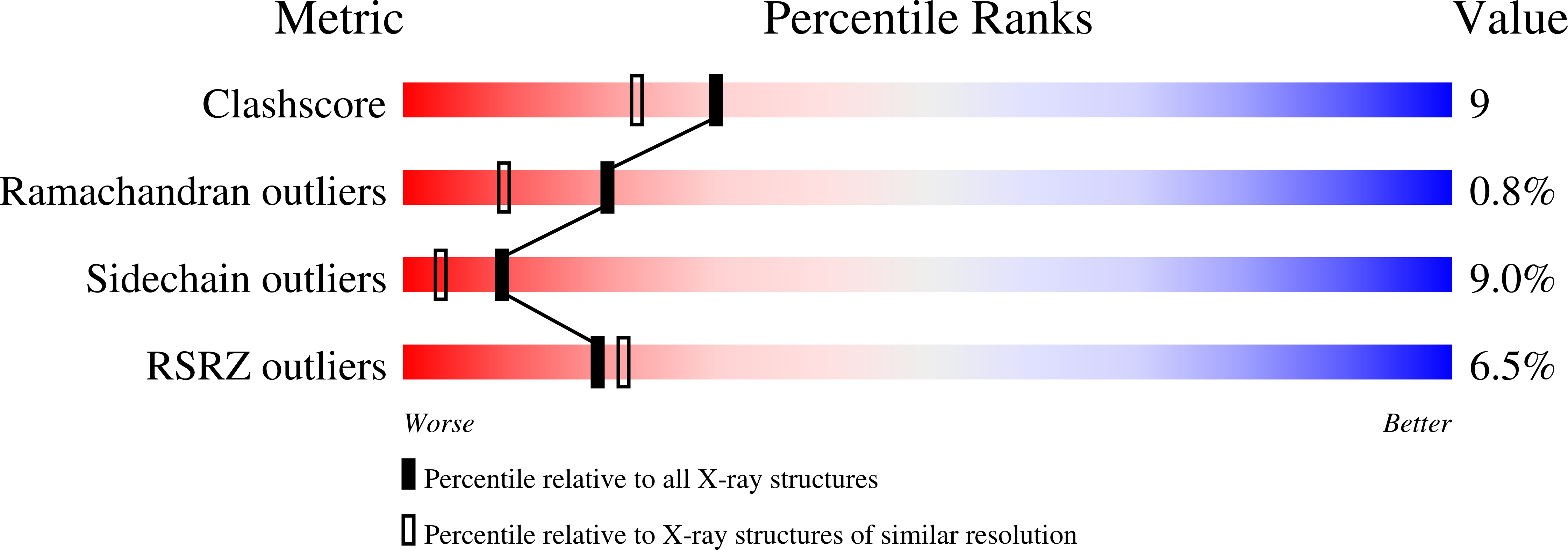Structure of the complex of bovine pancreatic phospholipase A2 with a transition-state analogue.
Sekar, K., Kumar, A., Liu, X., Tsai, M.D., Gelb, M.H., Sundaralingam, M.(1998) Acta Crystallogr D Biol Crystallogr 54: 334-341
- PubMed: 9761900
- DOI: https://doi.org/10.1107/s090744499701247x
- Primary Citation of Related Structures:
1MKV - PubMed Abstract:
The 1.89 A resolution structure of the complex of bovine pancreatic phospholipase A2 (PLA2) with the transition-state analogue L-1-O-octyl-2-heptylphosphonyl-sn-glycero-3-phosphoethanolamine (TSA) has been determined. The crystal of the complex is trigonal, space group P3121, a = b = 46.58 and c = 102.91 A and isomorphous to the native recombinant wild type (WT). The structure was refined to a final crystallographic R value of 18.0% including 957 protein atoms, 88 water molecules, one calcium ion and all 31 non-H atoms of the inhibitor at 1.89 A resolution. In all, 7 726 reflections [F>2sigma(F)] were used between 8.0 and 1.89 A resolution. The inhibitor is deeply locked into the active-site cleft and coordinates to the calcium ion by displacing the two water molecules in the calcium pentagonal bipyramid by the anionic O atoms of the phosphate and phosphonate group. The hydroxyl group of Tyr69 hydrogen bonds to the second anionic O atom of the phosphate group while that of the phosphonate group replaces the third water, 'catalytic' water, which forms a hydrogen bond to Ndelta1 of His48. The fourth water which also shares Ndelta1 of His48 is displaced by the steric hinderance of the inhibitor. The fifth conserved structural water is still present in the active site and forms a network of hydrogen bonds with the surrounding residues. The structure is compared to the other known TSA-PLA2 complexes.
Organizational Affiliation:
Biological Macromolecular Structure Center, Departments of Chemistry and Biochemistry, 100 West 18th Avenue, The Ohio State University, Columbus, OH 43210-1185, USA.