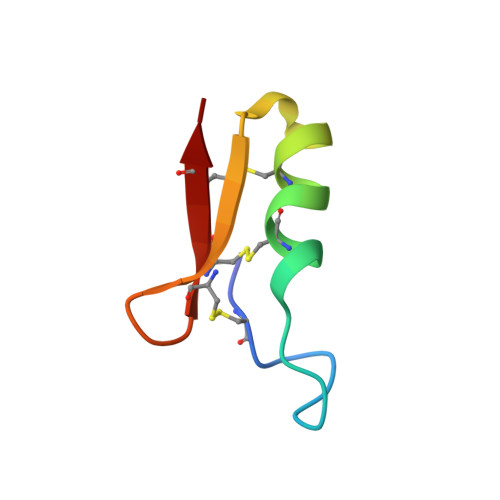
1H nuclear magnetic resonance study of the solution conformation of an antibacterial protein, sapecin.
Hanzawa, H., Shimada, I., Kuzuhara, T., Komano, H., Kohda, D., Inagaki, F., Natori, S., Arata, Y.(1990) FEBS Lett 269: 413-420
- PubMed: 2401368
- DOI: https://doi.org/10.1016/0014-5793(90)81206-4
- Primary Citation of Related Structures:
1L4V - PubMed Abstract:
The solution conformation of an antibacterial protein sapecin has been determined by 1H nuclear magnetic resonance (NMR) and dynamical simulated annealing calculations. It has been shown that the polypeptide fold consists of one flexible loop (residues 4-12), one helix (residues 15-23), and two extended strands (residues 24-31 and 34-40). It was found that the tertiary structure of sapecin is completely different from that of rabbit neutrophil defensin NP-5, which is homologous to sapecin in the amino acid sequences and also has the antibacterial activity. The three-dimensional structure determination has revealed that a basic-residue rich region and the hydrophobic surface face each other on the surface of sapecin.
Organizational Affiliation:
Faculty of Pharmaceutical Sciences, University of Tokyo, Japan.