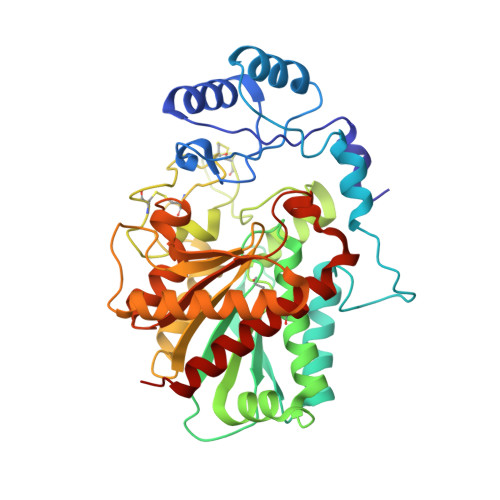
Human procarboxypeptidase B: three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI).
Barbosa Pereira, P.J., Segura-Martin, S., Oliva, B., Ferrer-Orta, C., Aviles, F.X., Coll, M., Gomis-Ruth, F.X., Vendrell, J.(2002) J Mol Biol 321: 537-547
- PubMed: 12162965
- DOI: https://doi.org/10.1016/s0022-2836(02)00648-4
- Primary Citation of Related Structures:
1KWM - PubMed Abstract:
Besides their classical role in alimentary protein degradation, zinc-dependant carboxypeptidases also participate in more selective regulatory processes like prohormone and neuropeptide processing or fibrinolysis inhibition in blood plasma. Human pancreatic procarboxypeptidase B (PCPB) is the prototype for those human exopeptidases that cleave off basic C-terminal residues and are secreted as inactive zymogens. One such protein is thrombin-activatable fibrinolysis inhibitor (TAFI), also known as plasma PCPB, which circulates in human plasma as a zymogen bound to plasminogen. The structure of human pancreatic PCPB displays a 95-residue pro-segment consisting of a globular region with an open-sandwich antiparallel-alpha antiparallel-beta topology and a C-terminal alpha-helix, which connects to the enzyme moiety. The latter is a 309-amino acid residue catalytic domain with alpha/beta hydrolase topology and a preformed active site, which is shielded by the globular domain of the pro-segment. The fold of the proenzyme is similar to previously reported procarboxypeptidase structures, also in that the most variable region is the connecting segment that links both globular moieties. However, the empty active site of human procarboxypeptidase B has two alternate conformations in one of the zinc-binding residues, which account for subtle differences in some of the key residues for substrate binding. The reported crystal structure, refined with data to 1.6A resolution, permits in the absence of an experimental structure, accurate homology modelling of TAFI, which may help to explain its properties.
Organizational Affiliation:
Institut de Biologia Molecular de Barcelona, C.I.D. - C.S.I.C., Jordi Girona, 18-26, E-08034, Barcelona, Spain.