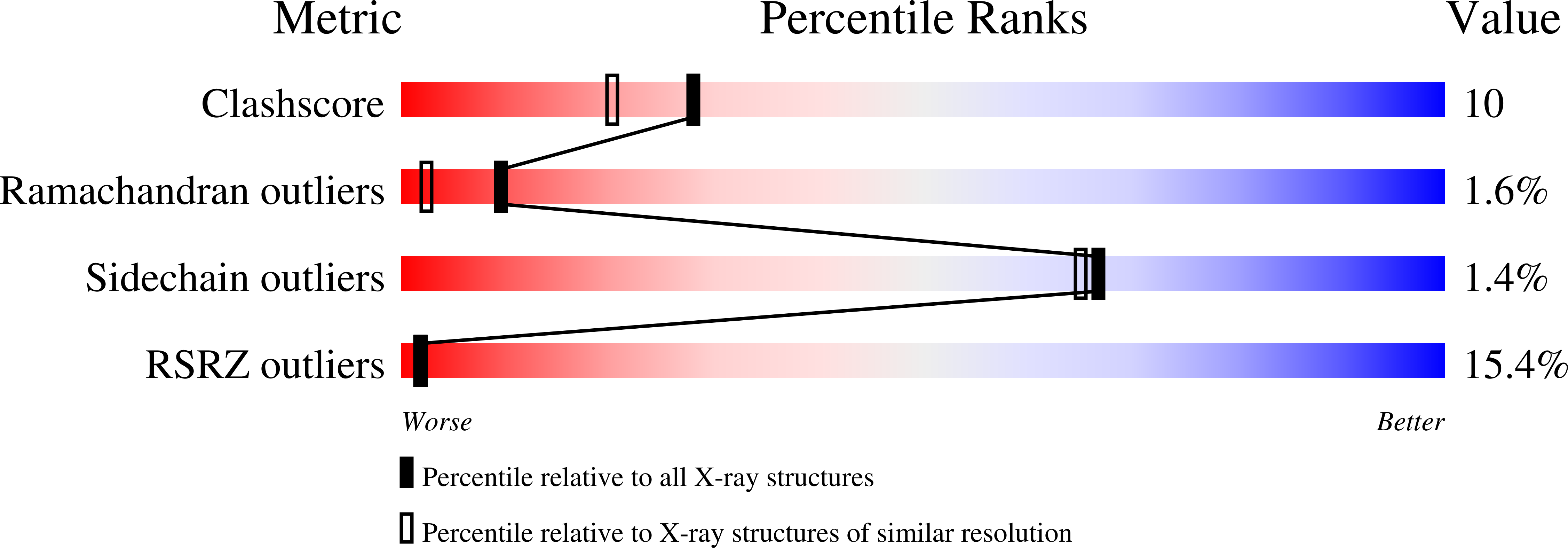
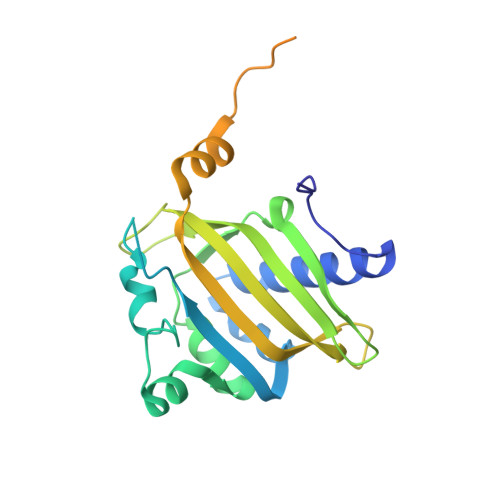
Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor.
Fribourg, S., Braun, I.C., Izaurralde, E., Conti, E.(2001) Mol Cell 8: 645-656
- PubMed: 11583626
- DOI: https://doi.org/10.1016/s1097-2765(01)00348-3
- Primary Citation of Related Structures:
1JKG, 1JN5 - PubMed Abstract:
TAP-p15 heterodimers have been implicated in the export of mRNAs through nuclear pore complexes (NPCs). We report a structural analysis of the interaction domains of TAP and p15 in a ternary complex with a Phe-Gly (FG) repeat of an NPC component. The TAP-p15 heterodimer is structurally similar to the homodimeric transport factor NTF2, but unlike NTF2, it is incompatible with either homodimerization or Ran binding. The NTF2-like heterodimer functions as a single structural unit in recognizing an FG repeat at a hydrophobic pocket present only on TAP and not on p15. This FG binding site interacts synergistically with a second site at the C terminus of TAP to mediate mRNA transport through the pore. In general, our findings suggest that FG repeats bind with a similar conformation to different classes of transport factors.
Organizational Affiliation:
European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117, Heidelberg, Germany