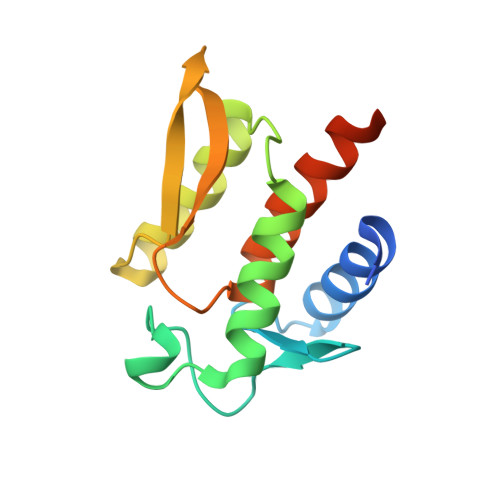
Structure of the N-terminal domain of Yersinia pestis YopH at 2.0 A resolution.
Evdokimov, A.G., Tropea, J.E., Routzahn, K.M., Copeland, T.D., Waugh, D.S.(2001) Acta Crystallogr D Biol Crystallogr 57: 793-799
- PubMed: 11375498
- DOI: https://doi.org/10.1107/s0907444901004875
- Primary Citation of Related Structures:
1HUF - PubMed Abstract:
Yersinia pestis, the causative agent of bubonic plague, injects effector proteins into the cytosol of mammalian cells that enable the bacterium to evade the immune response of the infected organism by interfering with eukaryotic signal transduction pathways. YopH is a modular effector composed of a C-terminal protein tyrosine phosphatase (PTPase) domain and a multifunctional N-terminal domain that not only orchestrates the secretion and translocation of YopH into eukaryotic cells but also binds tyrosine-phosphorylated target proteins to mediate substrate recognition. The crystal structure of the N-terminal domain of YopH (YopH(N); residues 1-130) has been determined at 2.0 A resolution. The amino-acid sequences that target YopH for secretion from the bacterium and translocation into eukaryotic cells form integral parts of this compactly folded domain. The structure of YopH(N) bears no resemblance to eukaryotic phosphotyrosine-binding domains, nor is it reminiscent of any known fold. Residues that have been implicated in phosphotyrosine-dependent protein binding are clustered together on one face of YopH(N), but the structure does not suggest a mechanism for protein-phosphotyrosine recognition.
Organizational Affiliation:
Protein Engineering Section, Macromolecular Crystallography Laboratory, National Cancer Institute at Frederick, PO Box B, Frederick, MD 21702-1201, USA.