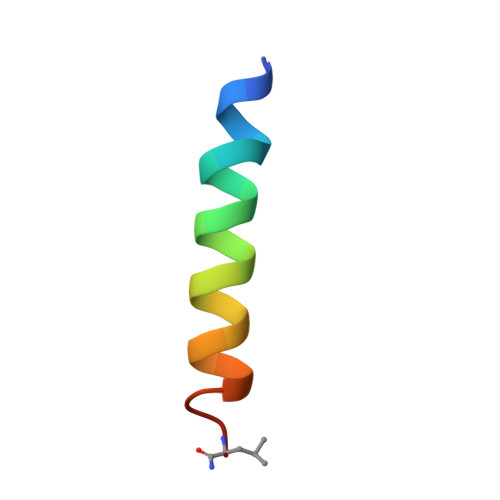
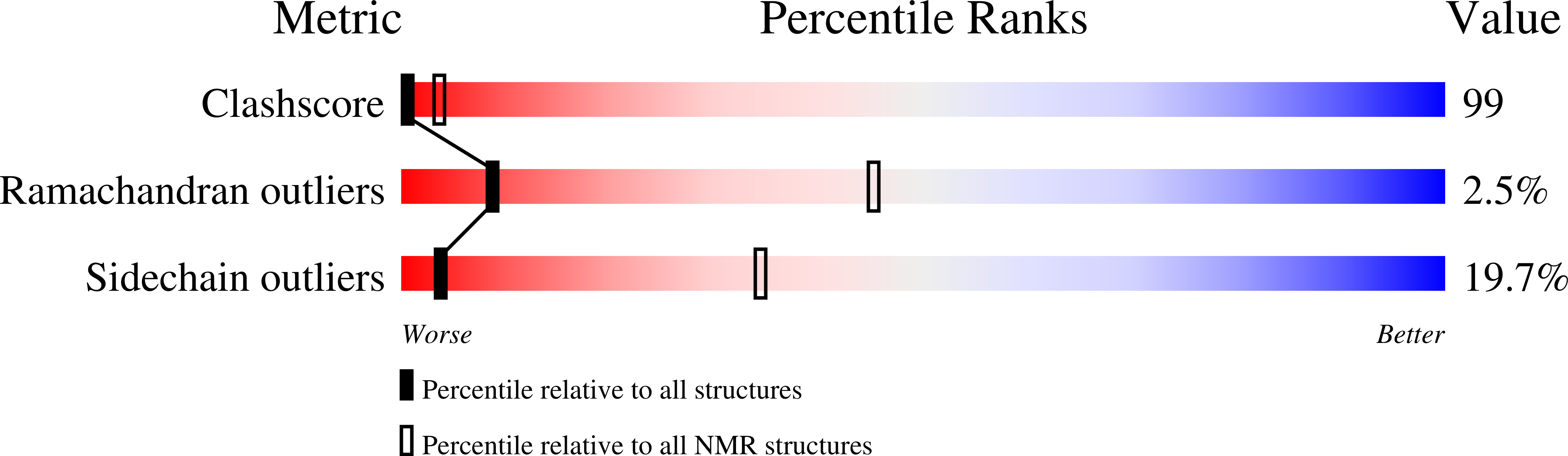Structural characterization of the antimicrobial peptide pleurocidin from winter flounder.
Syvitski, R.T., Burton, I., Mattatall, N.R., Douglas, S.E., Jakeman, D.L.(2005) Biochemistry 44: 7282-7293
- PubMed: 15882067
- DOI: https://doi.org/10.1021/bi0504005
- Primary Citation of Related Structures:
1Z64 - PubMed Abstract:
Pleurocidin is an antimicrobial peptide that was isolated from the mucus membranes of winter flounder (Pseudopleuronectes americanus) and contributes to the initial stages of defense against bacterial infection. From NMR structural studies with the uniformly (15)N-labeled peptide, a structure of pleurocidin was determined to be in a random coil conformation in aqueous solution whereas it assumes an alpha-helical structure in TFE and in dodecylphosphocholine (DPC) micelles. From (15)N relaxation studies, the helix is a rigid structure in the membrane-mimicking environment. Strong NOESY cross-peaks from the pleurocidin to the aliphatic chain on DPC confirm that pleurocidin is contained within the DPC micelle and not associated with the surface of the micelle. From diffusion studies it was determined that each micelle contains at least two pleurocidin molecules.
Organizational Affiliation:
College of Pharmacy, 5968 College Street, Dalhousie University, Halifax, Nova Scotia, Canada B3H 3J5.