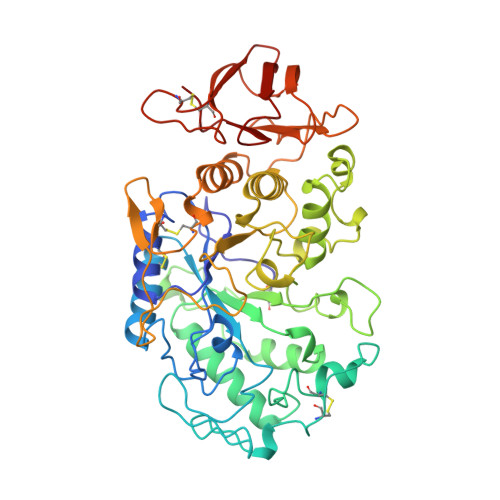
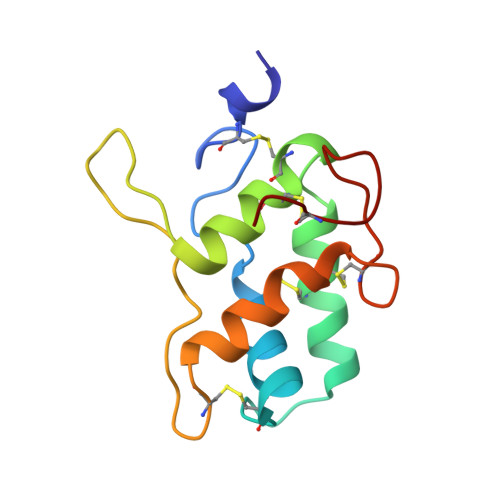
A novel strategy for inhibition of alpha-amylases: yellow meal worm alpha-amylase in complex with the Ragi bifunctional inhibitor at 2.5 A resolution.
Strobl, S., Maskos, K., Wiegand, G., Huber, R., Gomis-Ruth, F.X., Glockshuber, R.(1998) Structure 6: 911-921
- PubMed: 9687373
- DOI: https://doi.org/10.1016/s0969-2126(98)00092-6
- Primary Citation of Related Structures:
1TMQ - PubMed Abstract:
alpha-Amylases catalyze the hydrolysis of alpha-D-(1,4)-glucan linkages in starch and related compounds. There is a wide range of industrial and medical applications for these enzymes and their inhibitors. The Ragi bifunctional alpha-amylase/trypsin inhibitor (RBI) is the prototype of the cereal inhibitor superfamily and is the only member of this family that inhibits both trypsin and alpha-amylases. The mode of inhibition of alpha-amylases by these cereal inhibitors has so far been unknown. The crystal structure of yellow meal worm alpha-amylase (TMA) in complex with RBI was determined at 2.5 A resolution. RBI almost completely fills the substrate-binding site of TMA. Specifically, the free N terminus and the first residue (Ser1) of RBI interact with all three acidic residues of the active site of TMA (Asp185, Glu222 and Asp287). The complex is further stabilized by extensive interactions between the enzyme and inhibitor. Although there is no significant structural reorientation in TMA upon inhibitor binding, the N-terminal segment of RBI, which is highly flexible in the free inhibitor, adopts a 3(10)-helical conformation in the complex. RBI's trypsin-binding loop is located opposite the alpha-amylase-binding site, allowing simultaneous binding of alpha-amylase and trypsin. The binding of RBI to TMA constitutes a new inhibition mechanism for alpha-amylases and should be general for all alpha-amylase inhibitors of the cereal inhibitor superfamily. Because RBI inhibits two important digestive enzymes of animals, it constitutes an efficient plant defense protein and may be used to protect crop plants from predatory insects.
Organizational Affiliation:
Institut für Molekularbiologie und Biophysik, Eidgenössische Technische Hochschule Hönggerberg, Zürich, Switzerland.