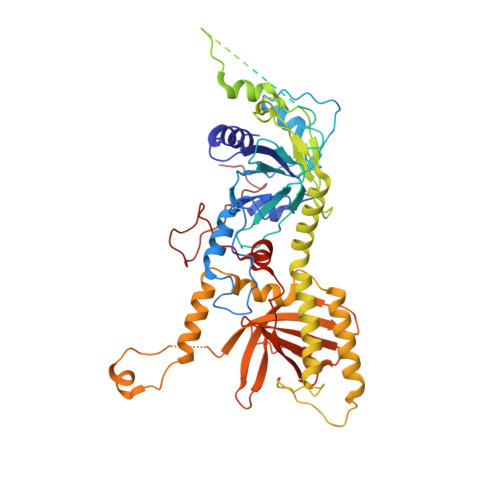
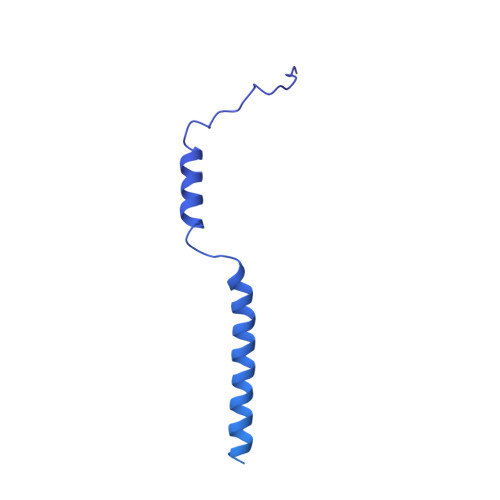
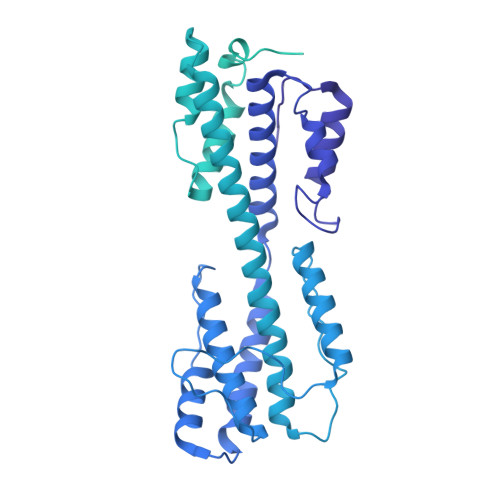
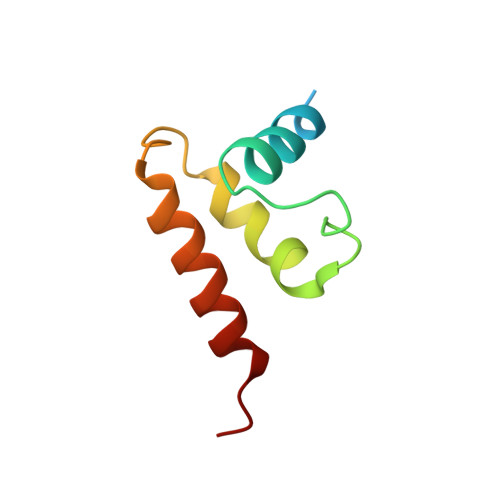
Structural basis for genome packaging, retention, and ejection in human cytomegalovirus.
Li, Z., Pang, J., Dong, L., Yu, X.(2021) Nat Commun 12: 4538-4538
- PubMed: 34315863
- DOI: https://doi.org/10.1038/s41467-021-24820-3
- Primary Citation of Related Structures:
7ET2, 7ET3, 7ETJ, 7ETM, 7ETO - PubMed Abstract:
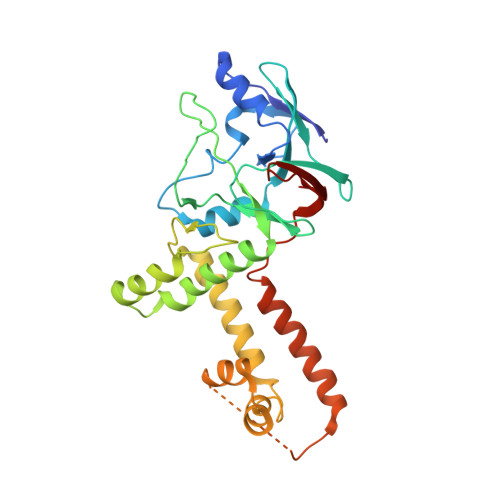
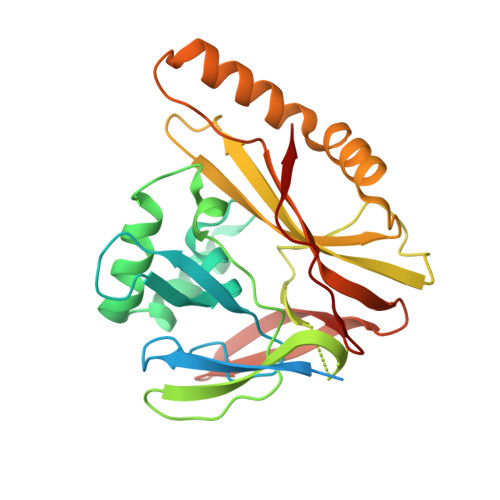
How the human cytomegalovirus (HCMV) genome-the largest among human herpesviruses-is packaged, retained, and ejected remains unclear. We present the in situ structures of the symmetry-mismatched portal and the capsid vertex-specific components (CVSCs) of HCMV. The 5-fold symmetric 10-helix anchor-uncommon among known portals-contacts the portal-encircling DNA, which is presumed to squeeze the portal as the genome packaging proceeds. We surmise that the 10-helix anchor dampens this action to delay the portal reaching a "head-full" packaging state, thus facilitating the large genome to be packaged. The 6-fold symmetric turret, latched via a coiled coil to a helix from a major capsid protein, supports the portal to retain the packaged genome. CVSCs at the penton vertices-presumed to increase inner capsid pressure-display a low stoichiometry, which would aid genome retention. We also demonstrate that the portal and capsid undergo conformational changes to facilitate genome ejection after viral cell entry.
Organizational Affiliation:
Cryo-Electron Microscopy Research Center, the CAS Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.