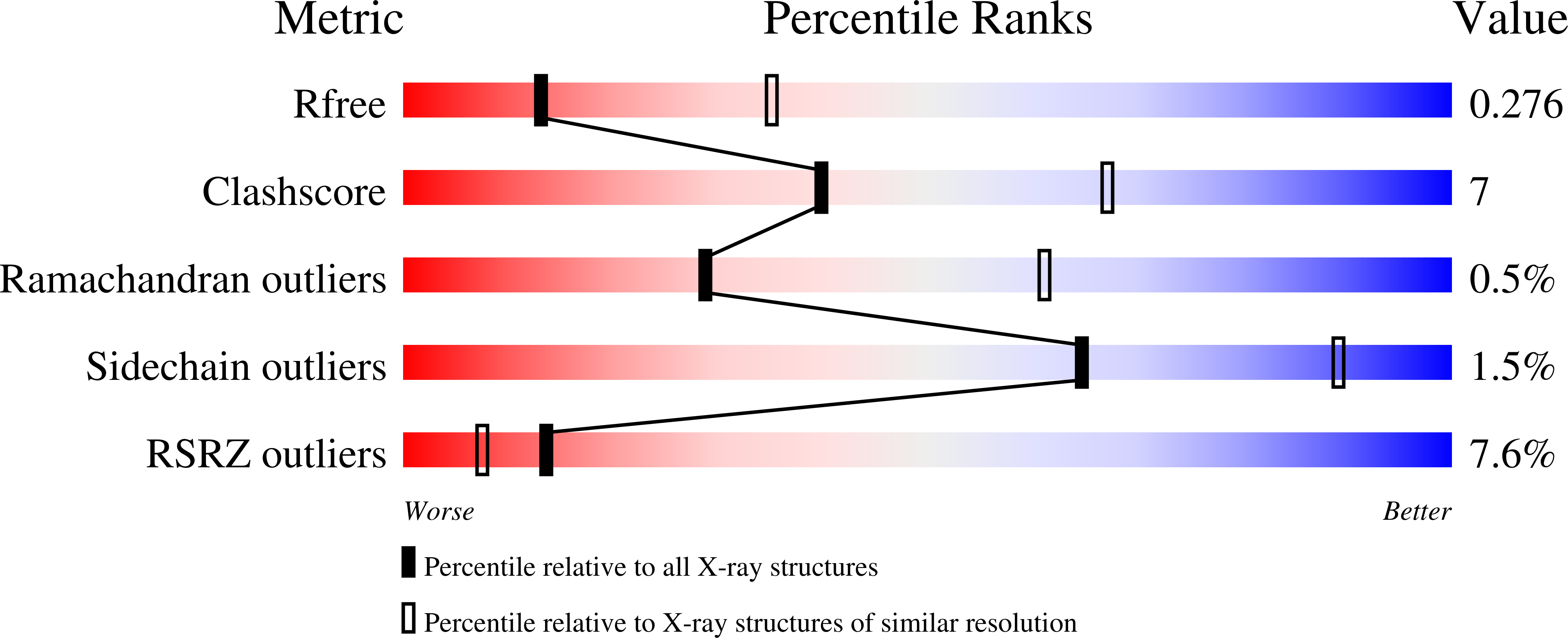
Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1.
Ferraro, D.J., Brown, E.N., Yu, C.L., Parales, R.E., Gibson, D.T., Ramaswamy, S.(2007) BMC Struct Biol 7: 10-10
- PubMed: 17349044
- DOI: https://doi.org/10.1186/1472-6807-7-10
- Primary Citation of Related Structures:
2GBW, 2GBX, 2I7F - PubMed Abstract:
The initial step involved in oxidative hydroxylation of monoaromatic and polyaromatic compounds by the microorganism Sphingobium yanoikuyae strain B1 (B1), previously known as Sphingomonas yanoikuyae strain B1 and Beijerinckia sp. strain B1, is performed by a set of multiple terminal Rieske non-heme iron oxygenases. These enzymes share a single electron donor system consisting of a reductase and a ferredoxin (BPDO-FB1). One of the terminal Rieske oxygenases, biphenyl 2,3-dioxygenase (BPDO-OB1), is responsible for B1's ability to dihydroxylate large aromatic compounds, such as chrysene and benzo[a]pyrene.
Organizational Affiliation:
Department of Biochemistry, University of Iowa Roy J. and Lucille A. Carver College of Medicine, Iowa City, Iowa, 52242, USA. daniel-ferraro@uiowa.edu