A conserved trimerization motif controls the topology of short coiled coils
Kammerer, R.A., Kostrewa, D., Progias, P., Honnappa, S., Avila, D., Lustig, A., Winkler, F.K., Pieters, J., Steinmetz, M.O.(2005) Proc Natl Acad Sci U S A 102: 13891-13896
- PubMed: 16172398
- DOI: https://doi.org/10.1073/pnas.0502390102
- Primary Citation of Related Structures:
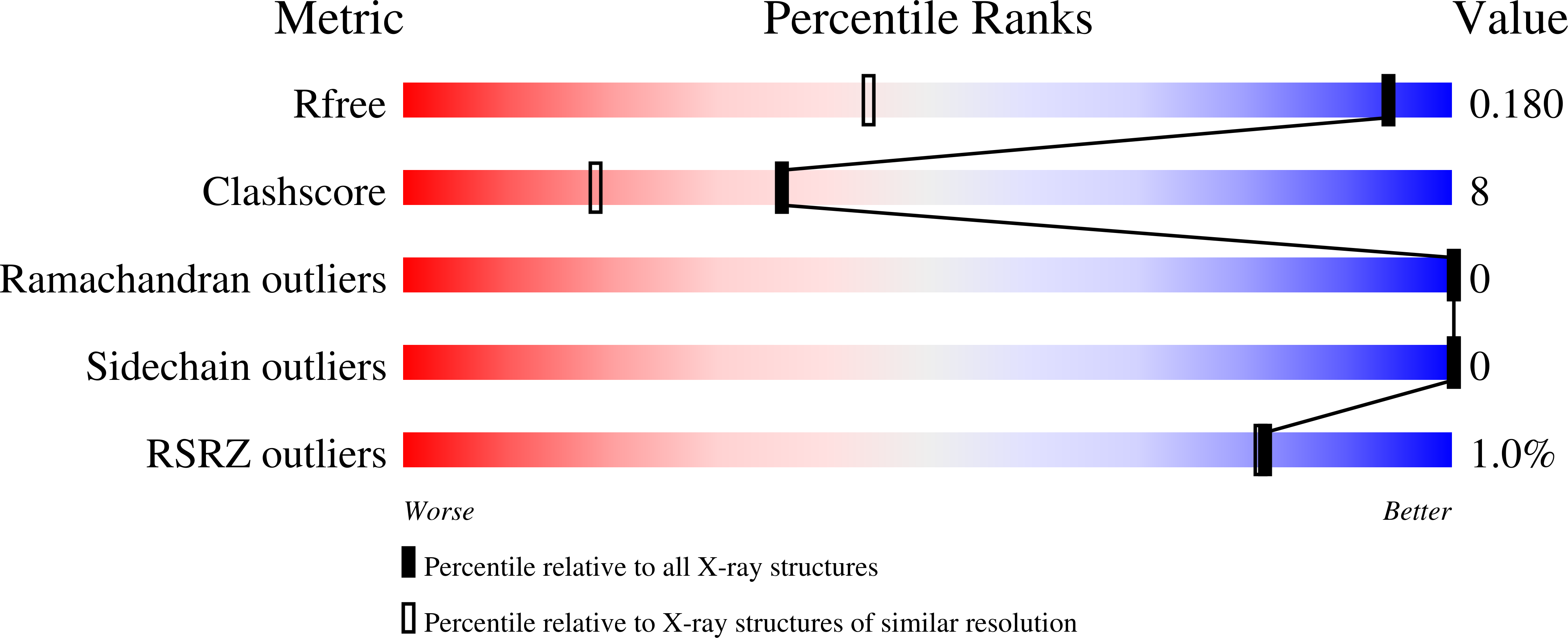
2AKF - PubMed Abstract:
In recent years, short coiled coils have been used for applications ranging from biomaterial to medical sciences. For many of these applications knowledge of the factors that control the topology of the engineered protein systems is essential. Here, we demonstrate that trimerization of short coiled coils is determined by a distinct structural motif that encompasses specific networks of surface salt bridges and optimal hydrophobic packing interactions. The motif is conserved among intracellular, extracellular, viral, and synthetic proteins and defines a universal molecular determinant for trimer formation of short coiled coils. In addition to being of particular interest for the biotechnological production of candidate therapeutic proteins, these findings may be of interest for viral drug development strategies.
Organizational Affiliation:
Wellcome Trust Centre for Cell-Matrix Research, Faculty of Life Sciences, University of Manchester, Manchester M13 PT, United Kingdom.