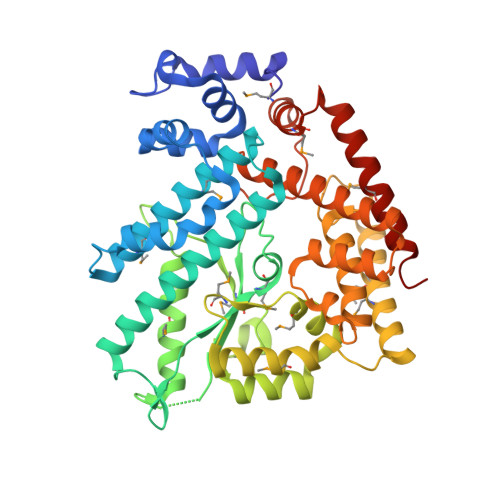
Structure of the n-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase
Xu, Y., Zhang, R., Joachimiak, A., Carr, P.D., Huber, T., Vasudevan, S.G., Ollis, D.L.(2004) Structure 12: 861-869
- PubMed: 15130478
- DOI: https://doi.org/10.1016/j.str.2004.02.029
- Primary Citation of Related Structures:
1V4A - PubMed Abstract:
We report the crystal structure of the N-terminal domain of Escherichia coli adenylyltransferase that catalyzes the reversible nucleotidylation of glutamine synthetase (GS), a key enzyme in nitrogen assimilation. This domain (AT-N440) catalyzes the deadenylylation and subsequent activation of GS. The structure has been divided into three subdomains, two of which bear some similarity to kanamycin nucleotidyltransferase (KNT). However, the orientation of the two domains in AT-N440 differs from that in KNT. The active site of AT-N440 has been identified on the basis of structural comparisons with KNT, DNA polymerase beta, and polyadenylate polymerase. AT-N440 has a cluster of metal binding residues that are conserved in polbeta-like nucleotidyl transferases. The location of residues conserved in all ATase sequences was found to cluster around the active site. Many of these residues are very likely to play a role in catalysis, substrate binding, or effector binding.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, James Cook University, Townsville, Queensland 4811, Australia. yibin.xu@jcu.edu.au