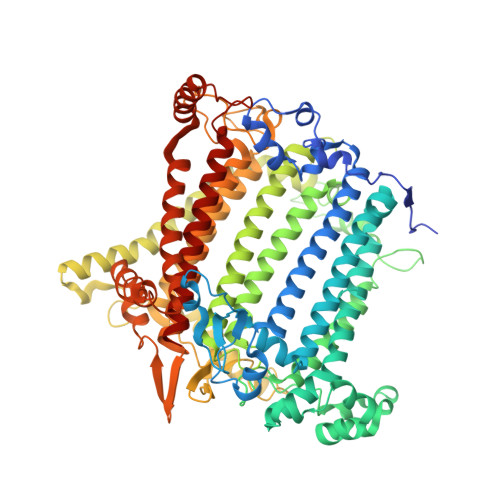
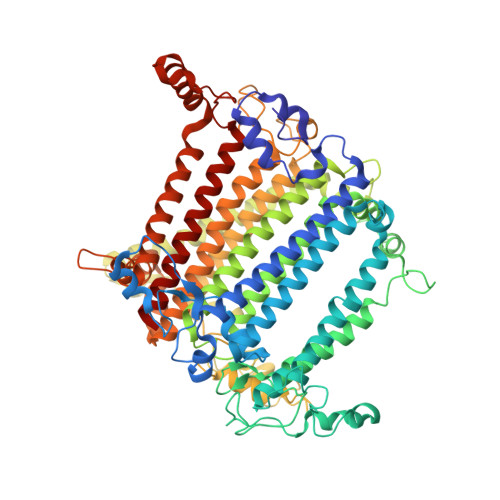
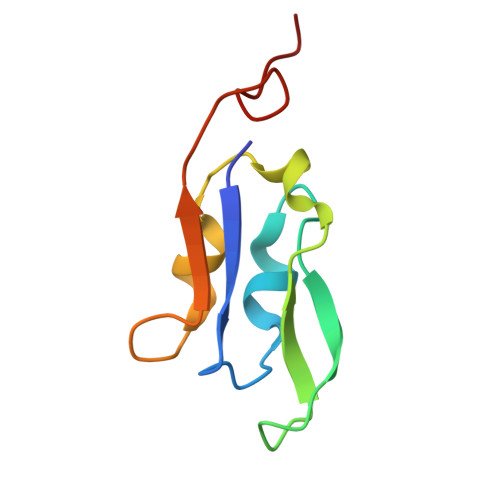
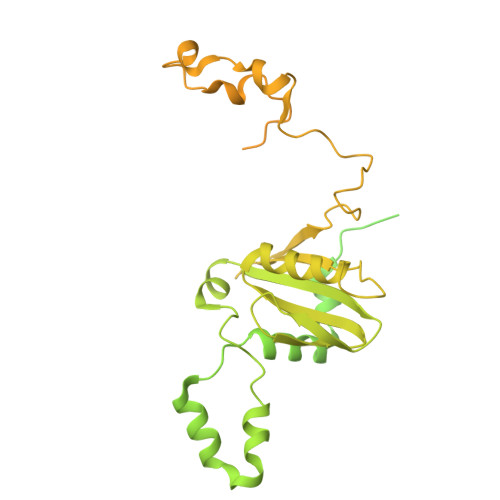
Structural insights into the divergent evolution of a photosystem I supercomplex in Euglena gracilis.
Kato, K., Nakajima, Y., Sakamoto, R., Kumazawa, M., Ifuku, K., Ishikawa, T., Shen, J.R., Takabayashi, A., Nagao, R.(2025) Sci Adv 11: eaea6241-eaea6241
- PubMed: 41171917
- DOI: https://doi.org/10.1126/sciadv.aea6241
- Primary Citation of Related Structures:
9VFJ - PubMed Abstract:
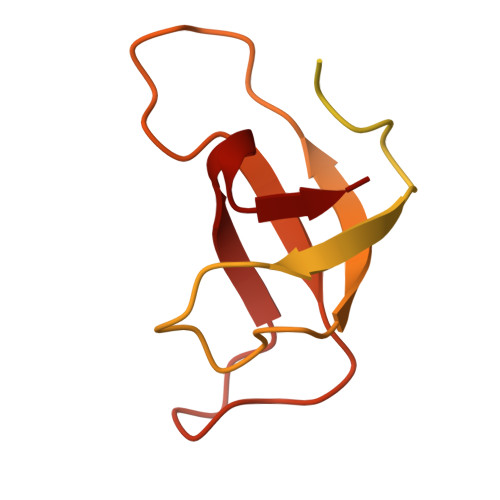
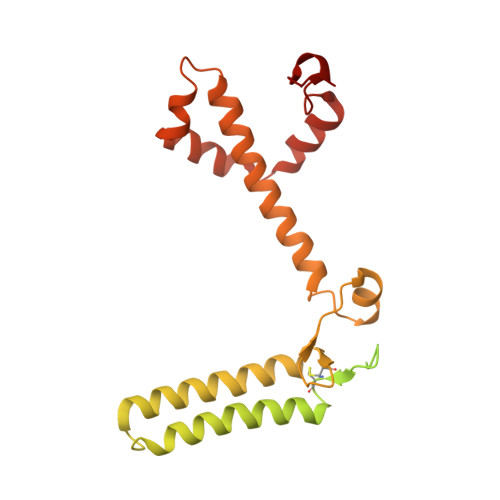
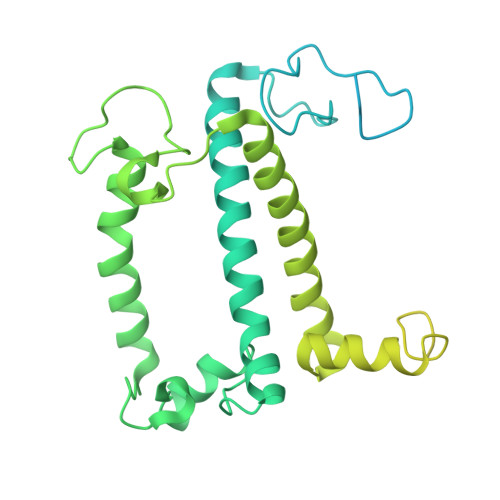
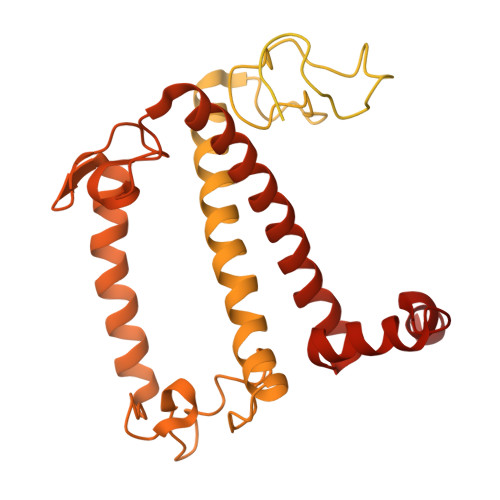
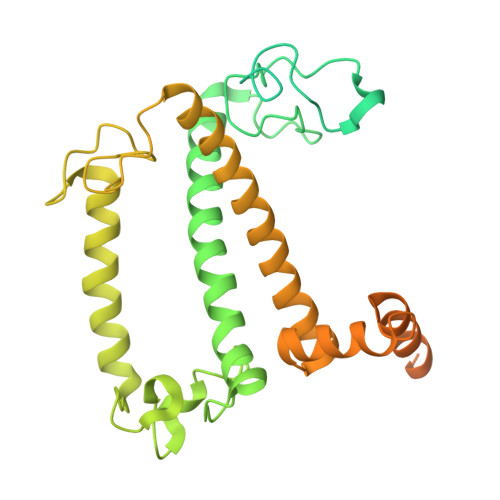
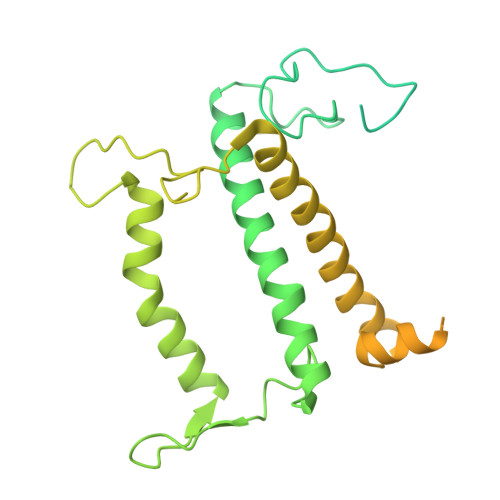
Photosystem I (PSI) forms supercomplexes with light-harvesting complexes (LHCs) to perform oxygenic photosynthesis. Here, we report a 2.82-angstrom cryo-electron microscopy structure of the PSI-LHCI supercomplex from Euglena gracilis , a eukaryotic alga with secondary green alga-derived plastids. The structure reveals a PSI monomer core with eight subunits and 13 asymmetrically arranged LHCI proteins. Euglena LHCIs bind diadinoxanthin, which is one of the carotenoids typically associated with red-lineage LHCs and is not present in the canonical LHCI belt found in green-lineage PSI-LHCI structures. Phylogenetic analysis shows that the Euglena LHCIs originated from LHCII-related clades rather than from the green-lineage LHCI group and that the nuclear-encoded PSI subunit PsaD likely originated from cyanobacteria via horizontal gene transfer. These observations indicate a mosaic origin of the Euglena PSI-LHCI. Our findings uncover a noncanonical light-harvesting architecture and highlight the structural and evolutionary plasticity of photosynthetic systems, illustrating how endosymbiotic acquisition and lineage-specific adaptation shape divergent light-harvesting strategies.
- Research Institute for Interdisciplinary Science, Advanced Research Field, and Graduate School of Environmental, Life, Natural Science and Technology, Okayama University, Okayama, Okayama 700-8530, Japan.
Organizational Affiliation: