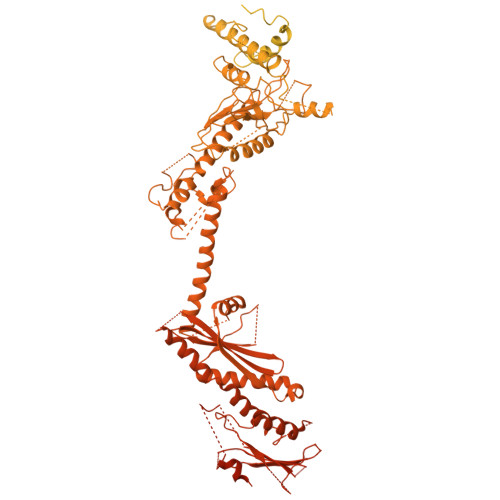
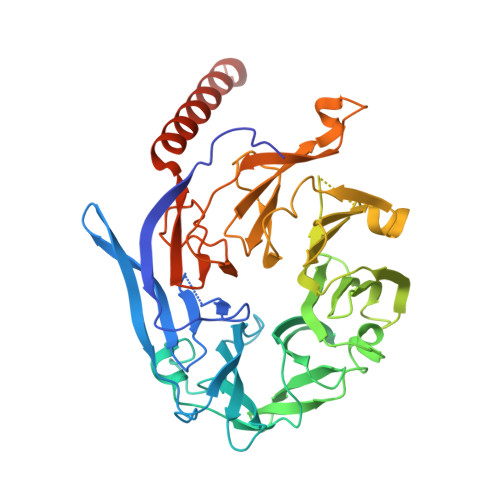
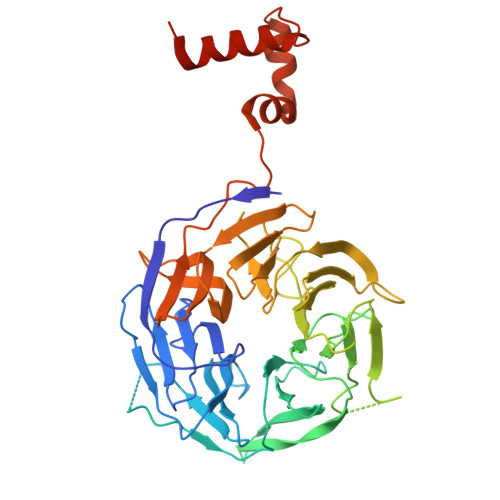Architecture of the human KICSTOR and GATOR1-KICSTOR complexes.
Teng, F., Zeng, H., Mai, X., Chen, S., Wang, L., Feng, Z., Tian, S., Wang, S., Stjepanovic, G., Lim, C.Y., Su, M.Y.(2025) Nat Struct Mol Biol 32: 2587-2600
- PubMed: 41198956
- DOI: https://doi.org/10.1038/s41594-025-01693-4
- Primary Citation of Related Structures:
9V0J, 9V6E, 9V80, 9V86, 9V9N, 9VAN - PubMed Abstract:
The human KICSTOR complex, comprising KPTN, ITFG2, C12orf66 and the scaffolding protein SZT2, anchors the mTORC1 inhibitor GATOR1 to lysosomes. Mutations affecting KICSTOR subunits are associated with severe neurodevelopmental and epileptic disorders. Loss of KICSTOR mimics GATOR1 inactivation, resulting in constitutive mTORC1 activation, highlighting its critical role in nutrient sensing. Here, we used cryo-electron microscopy and computational modeling to determine the architectures of KICSTOR and the GATOR1-KICSTOR supercomplex. We show that SZT2 forms a crescent-shaped scaffold with repetitive tandem units, binding the ITFG2-KPTN heterodimer and C12orf66 at its C terminus. Structural and biochemical analyses revealed that GATOR1 binds the SZT2 N-terminal domain through NPRL3; disruption of this interaction hyperactivates mTORC1 and mislocalizes TFE3 independently of nutrient status. We further demonstrate the membrane-binding ability of KICSTOR, with SZT2 and C12orf66 preferentially interacting with negatively charged lipids-a requirement for lysosomal localization. These findings identify how KICSTOR positions GATOR1 on lysosomes to regulate nutrient-dependent mTORC1 signaling.
- Department of Biochemistry, Key University Laboratory of Metabolism and Health of Guangdong, SUSTech Homeostatic Medicine Institute, School of Medicine, Southern University of Science and Technology, Shenzhen, China.
Organizational Affiliation: